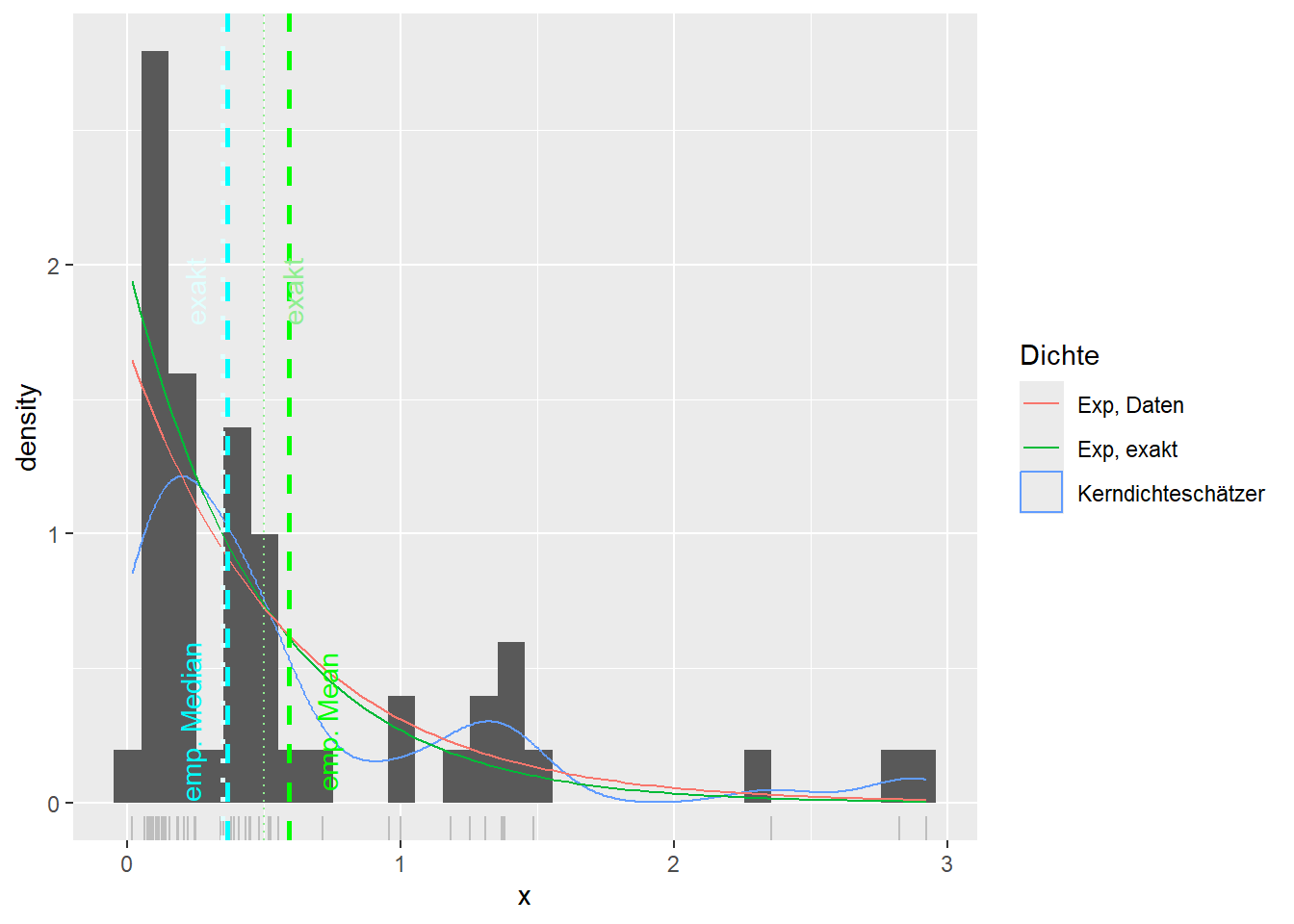
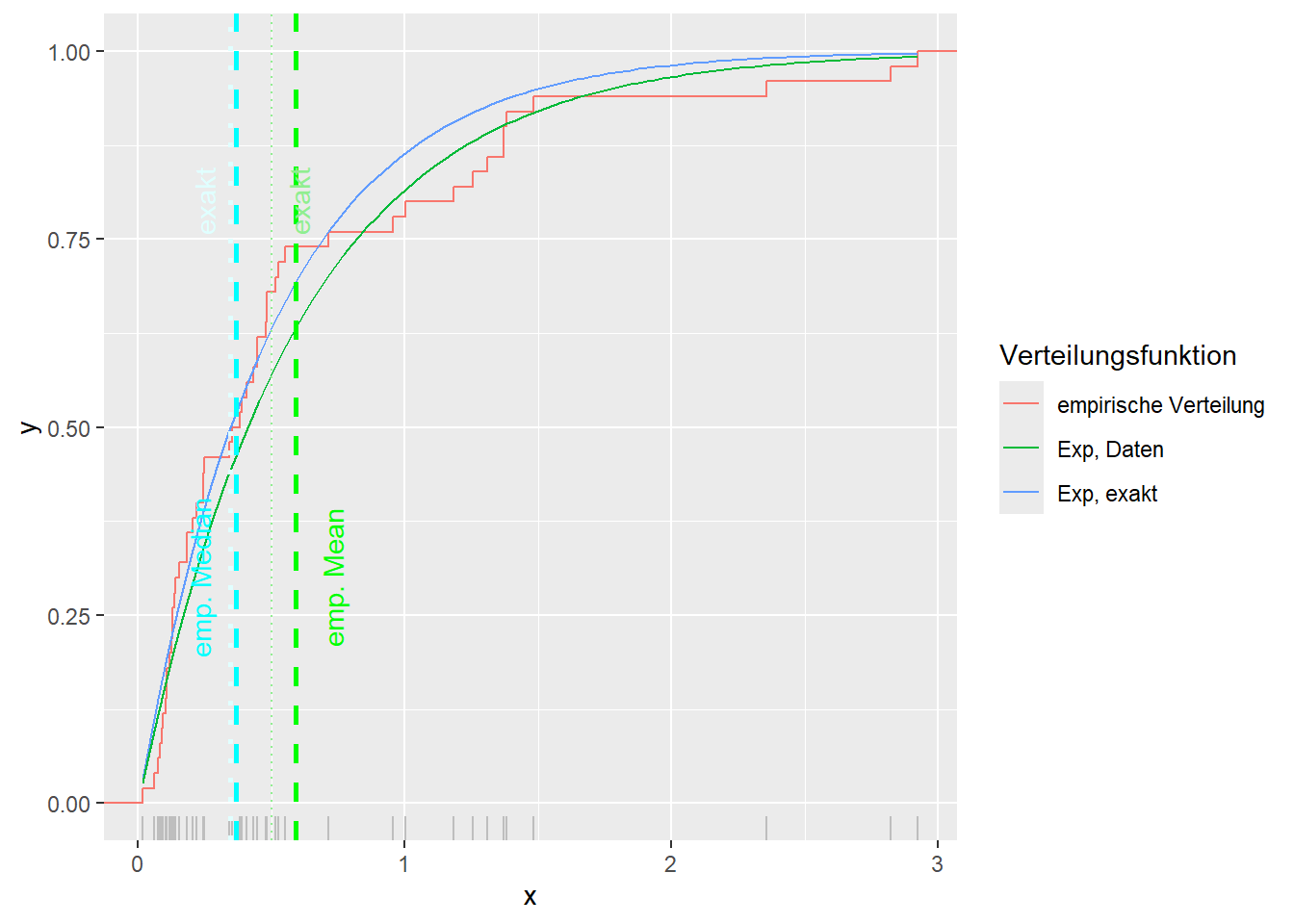
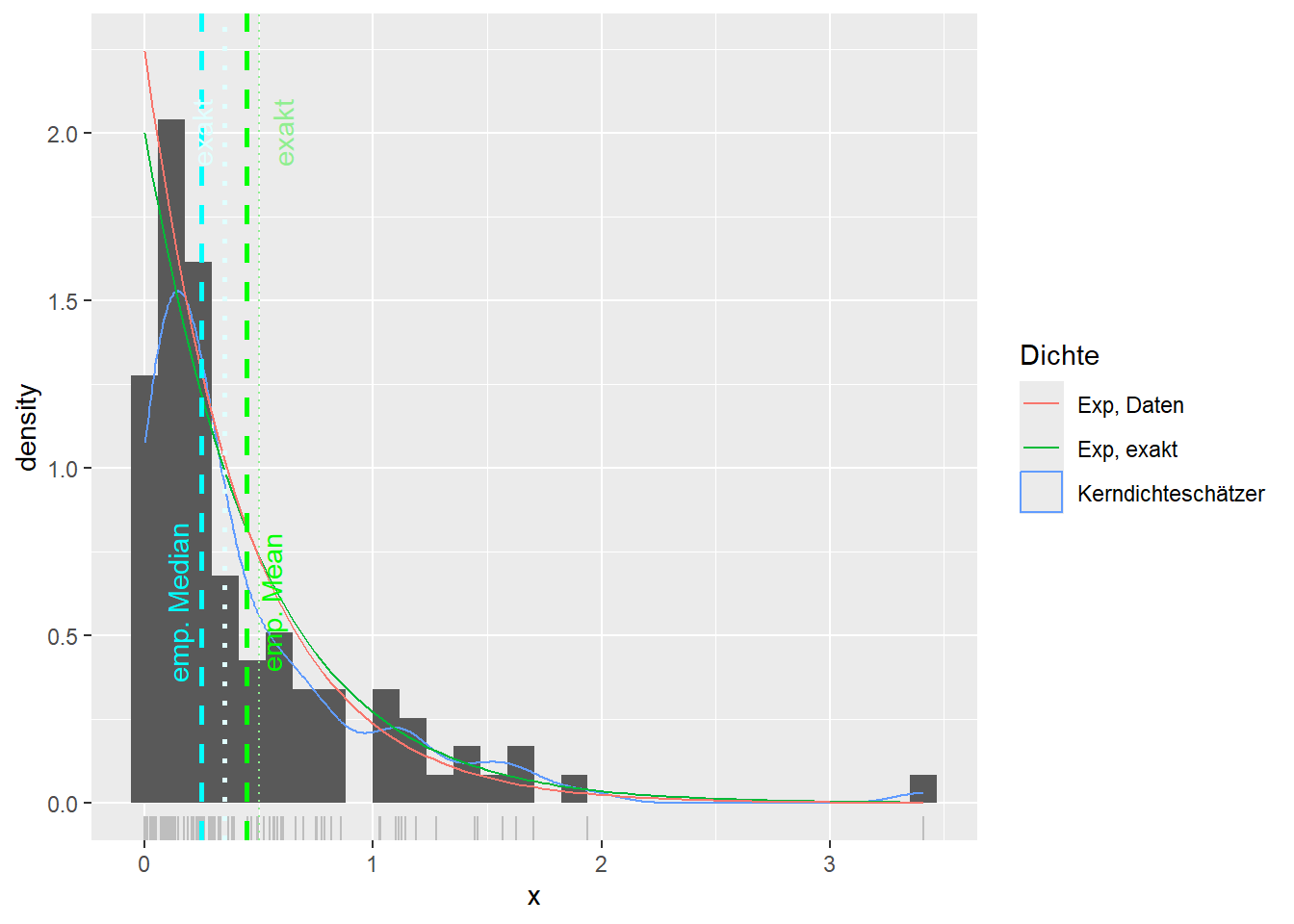
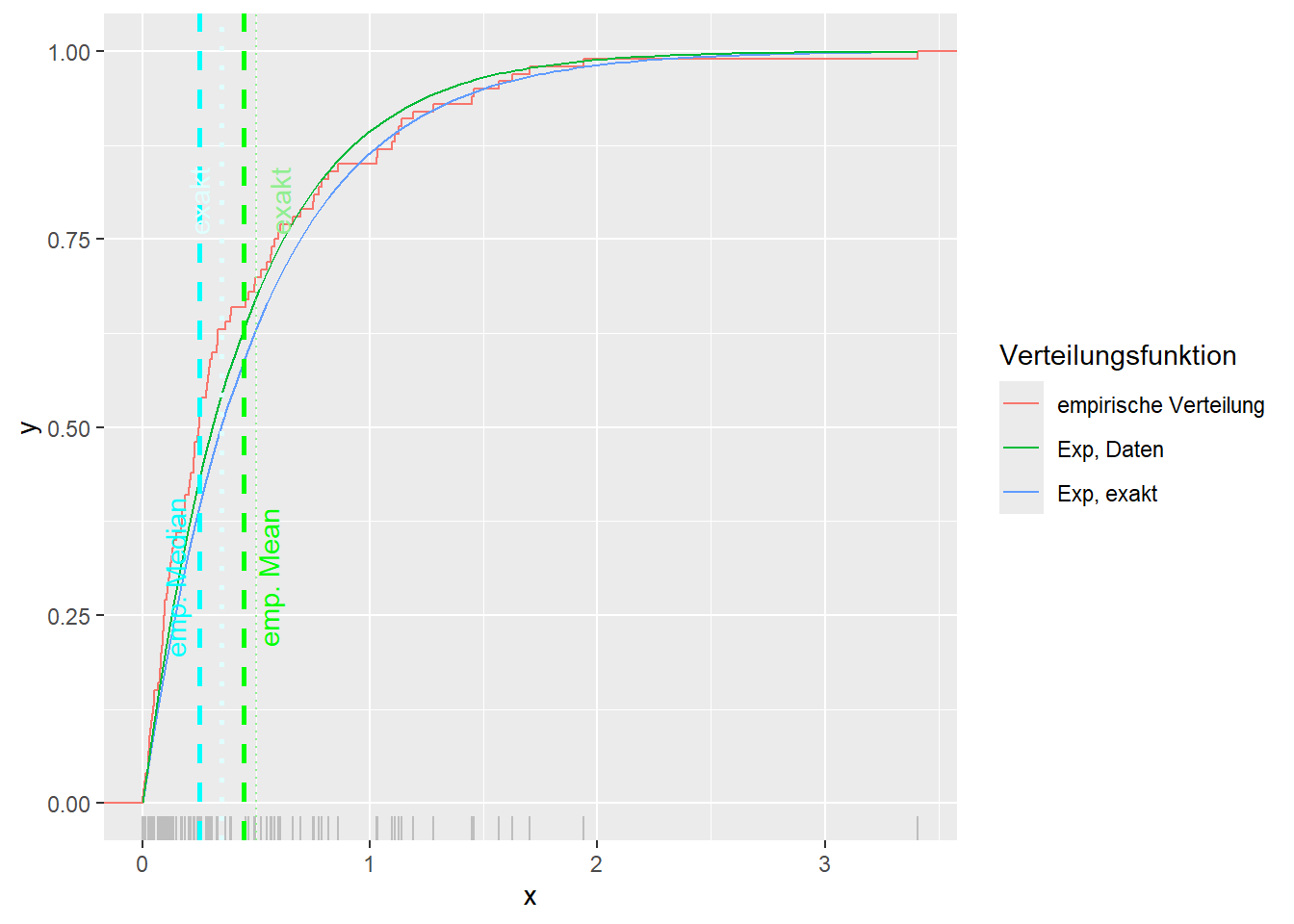
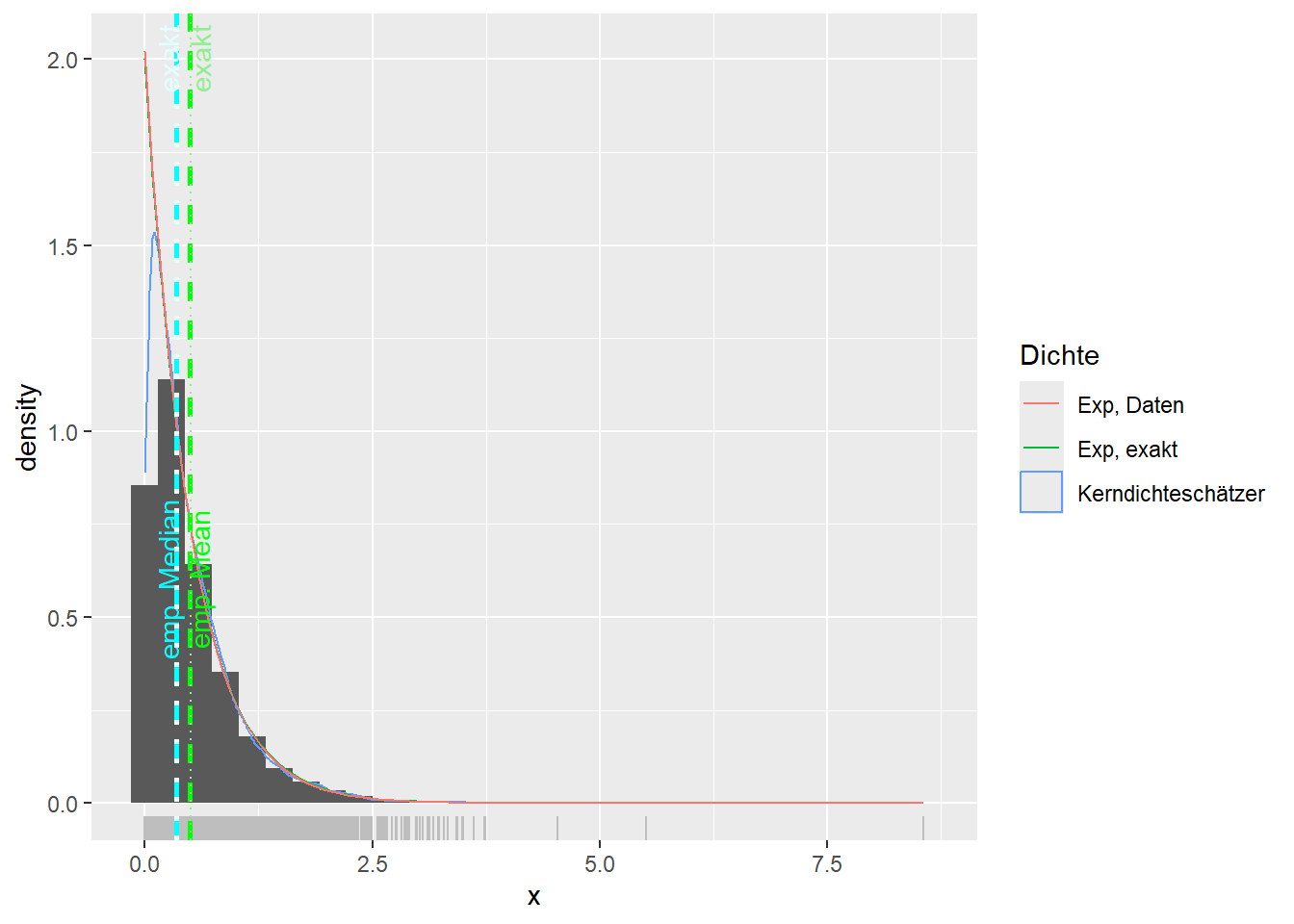
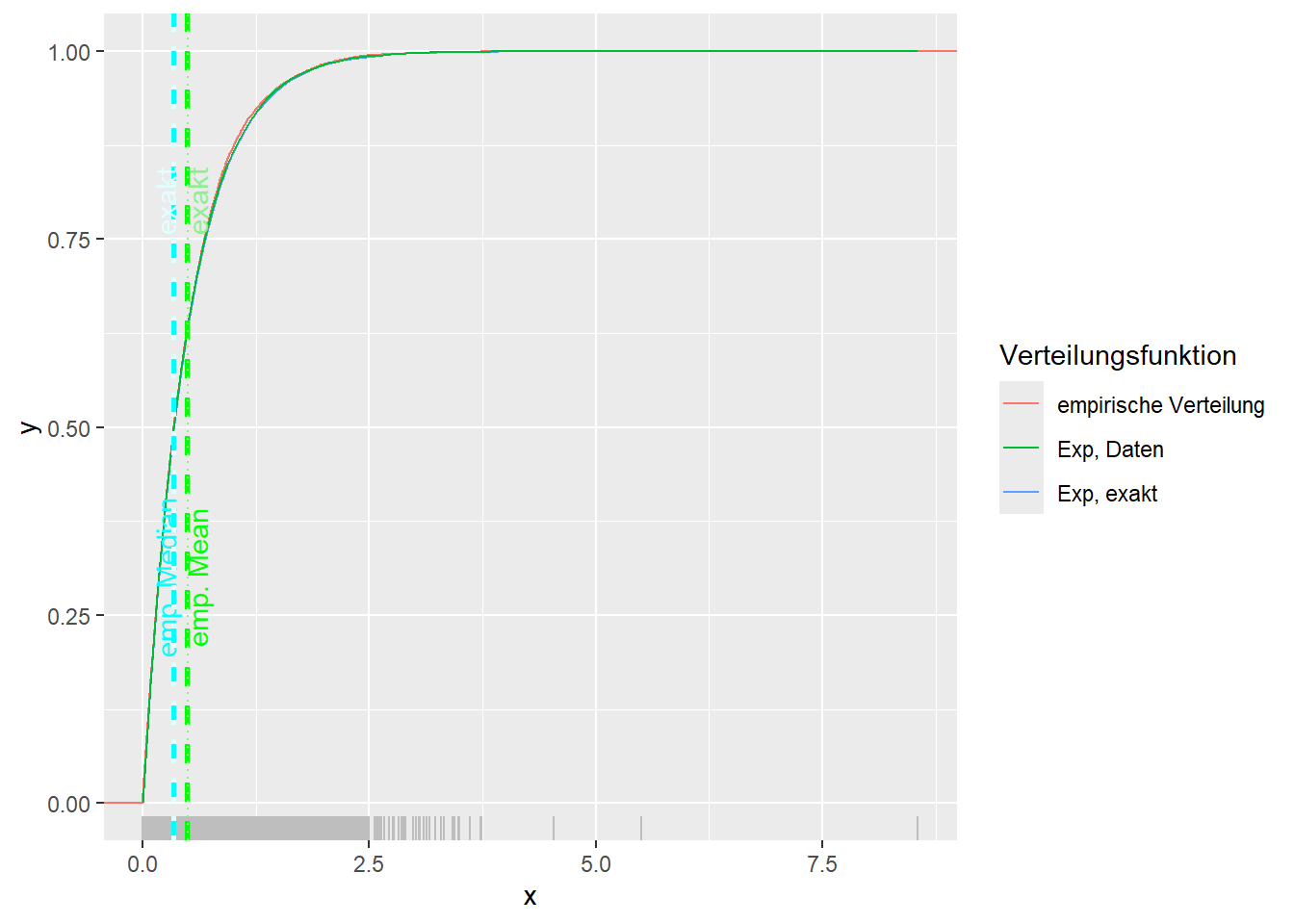
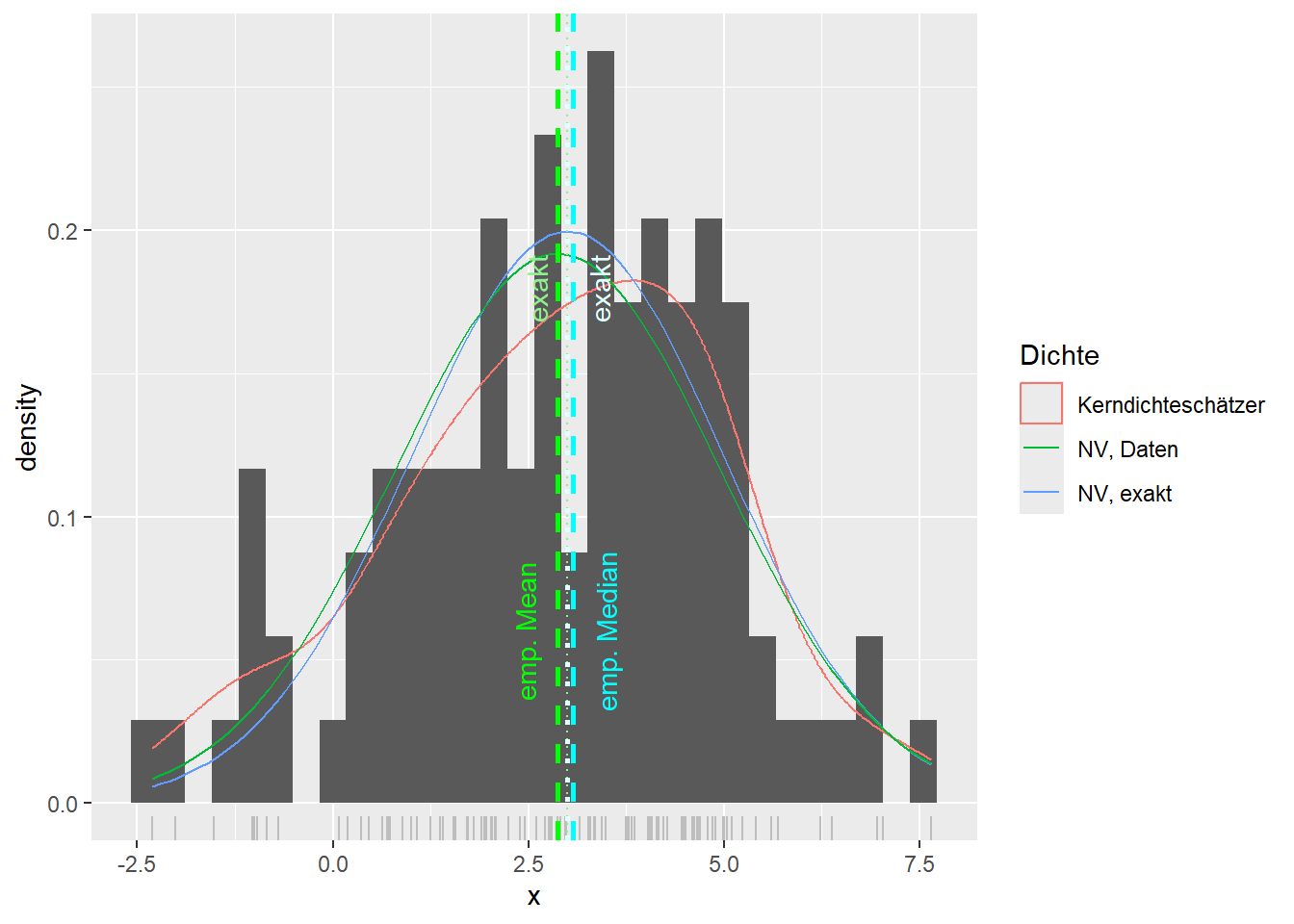
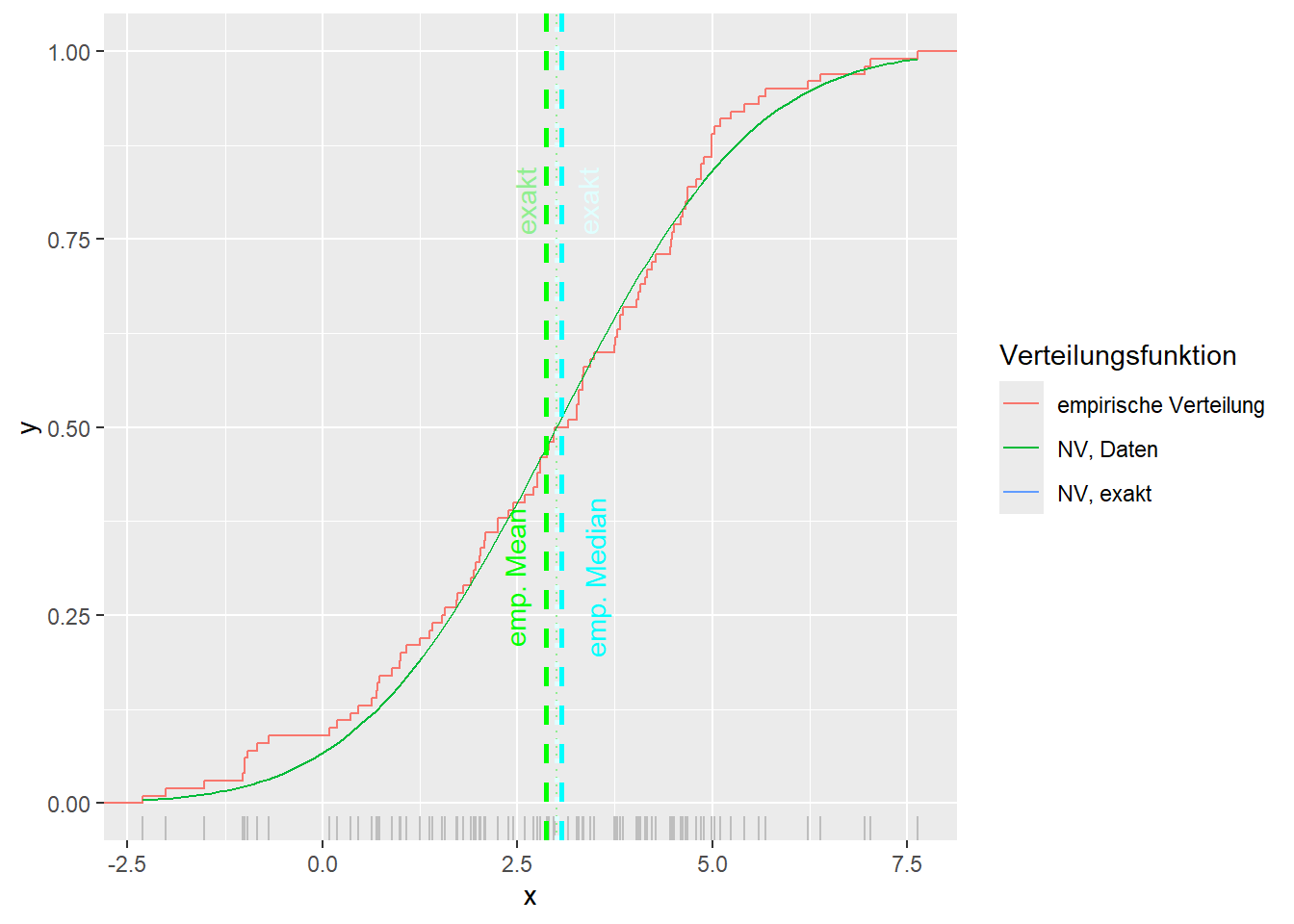
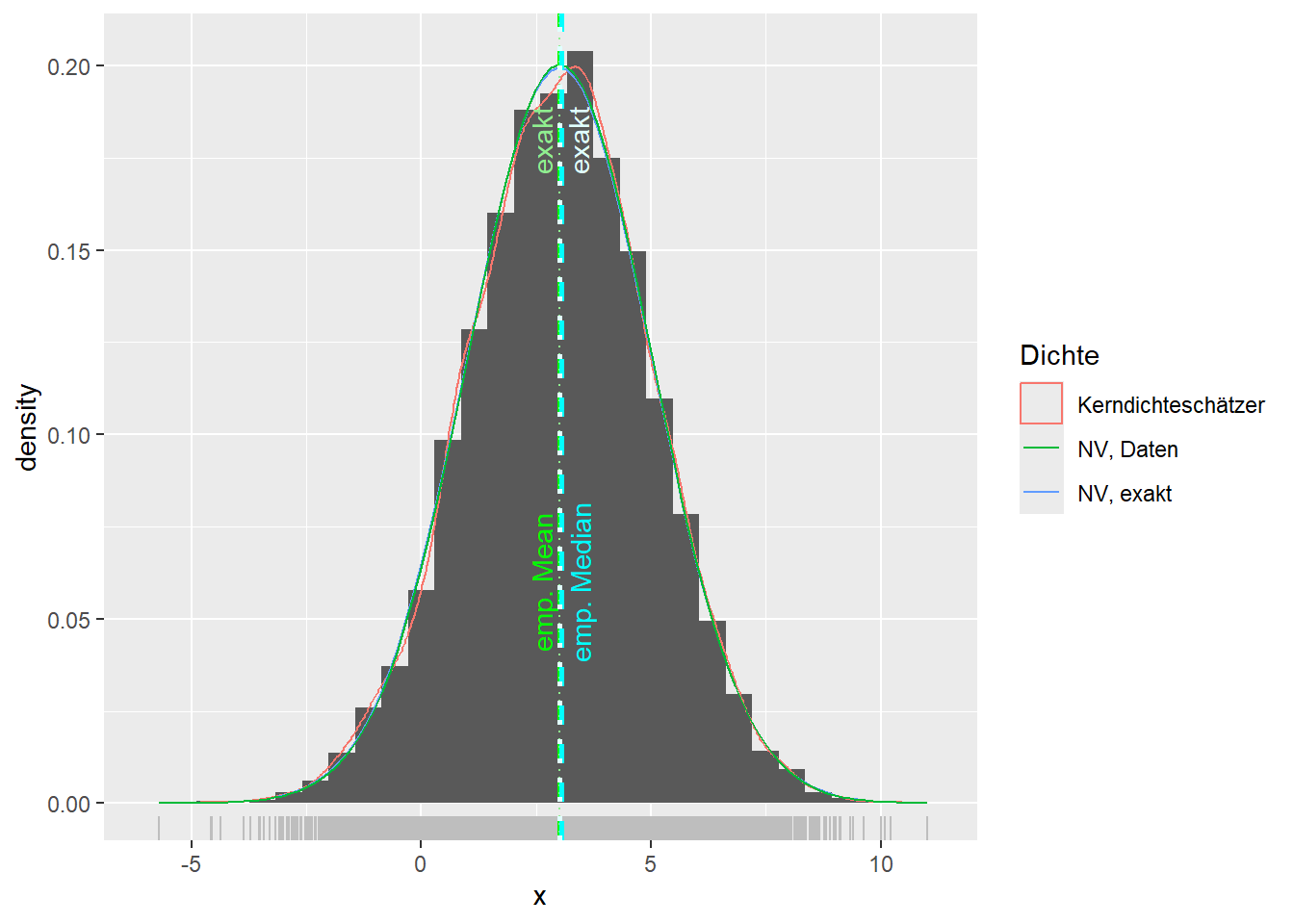
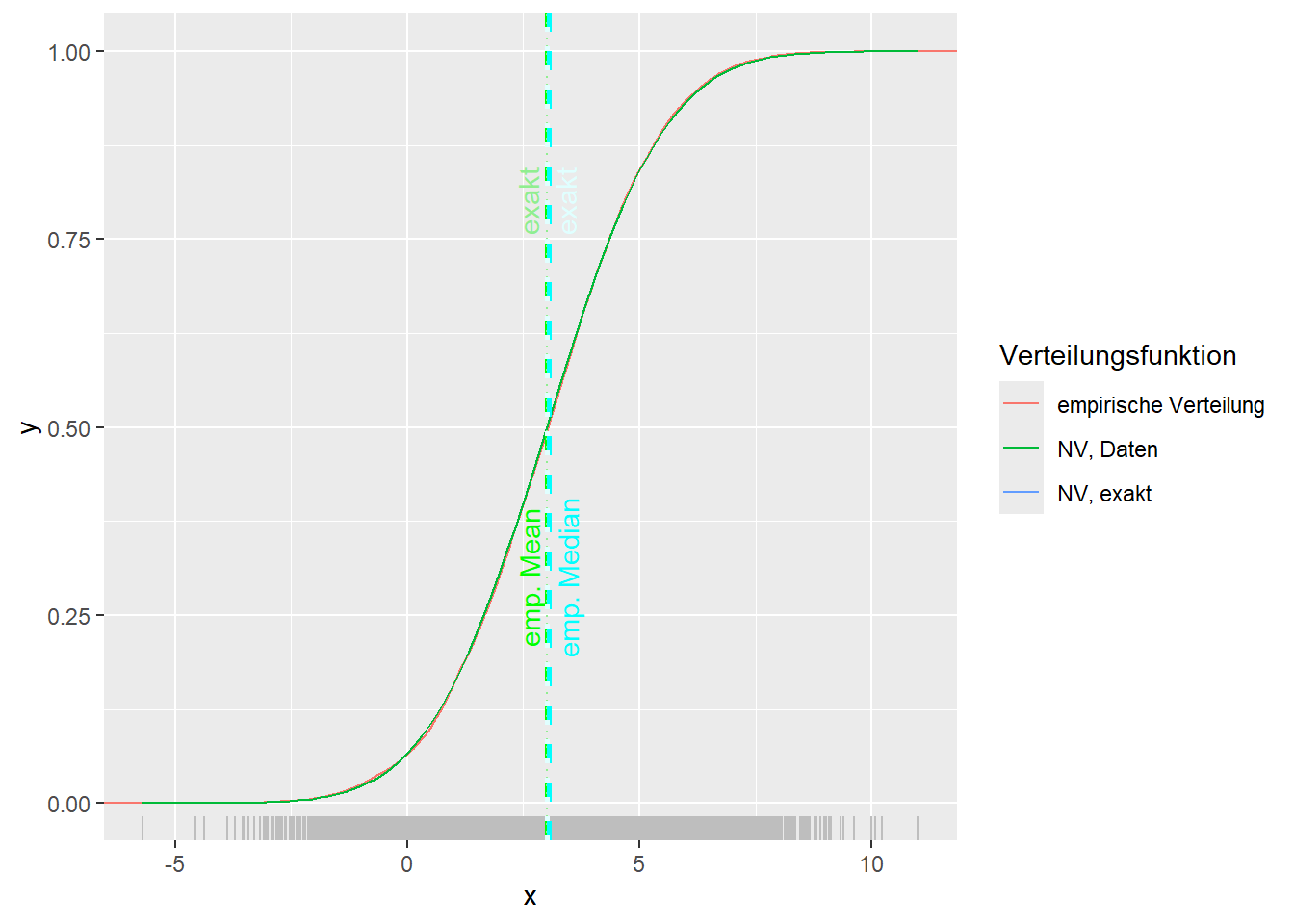
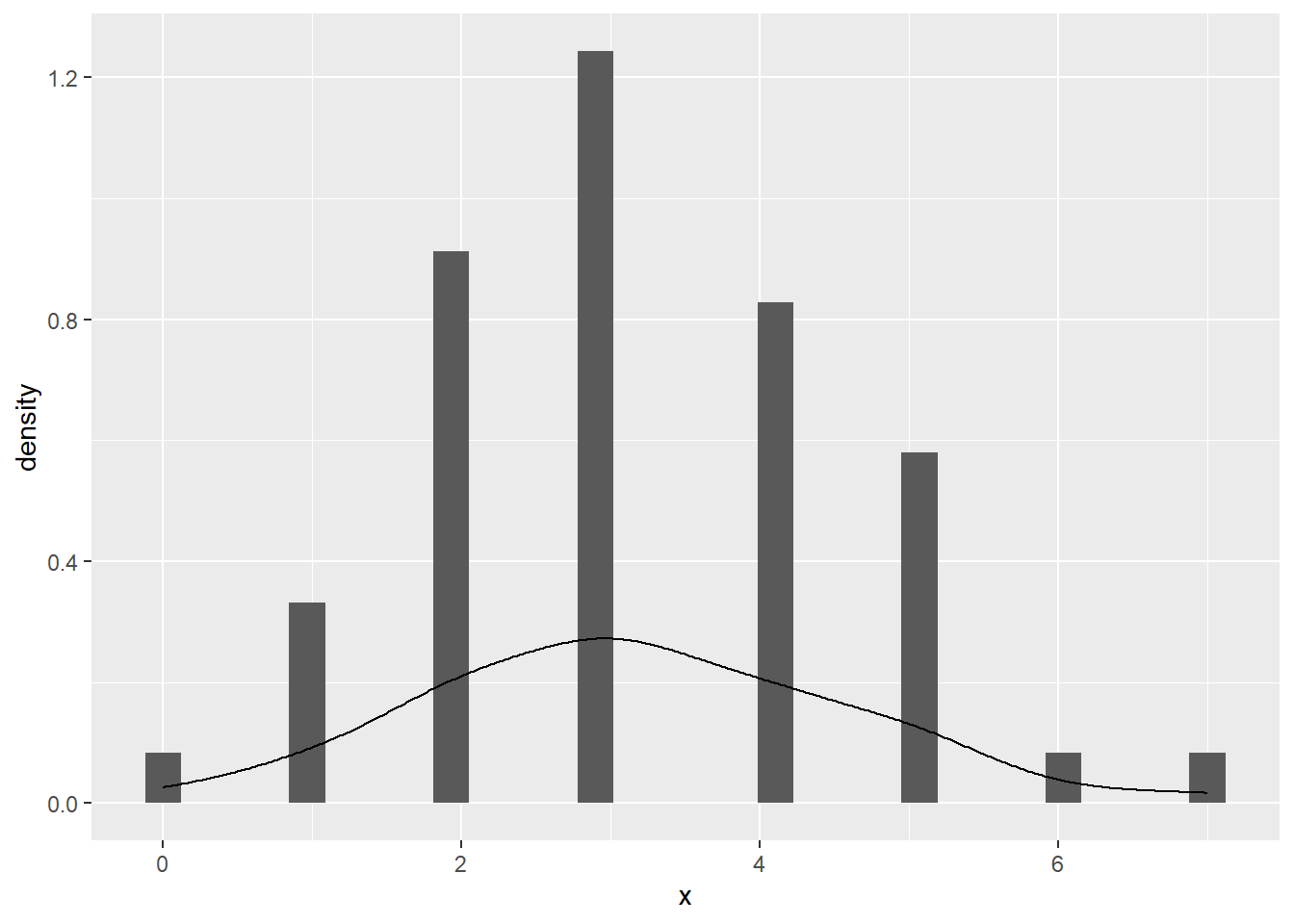
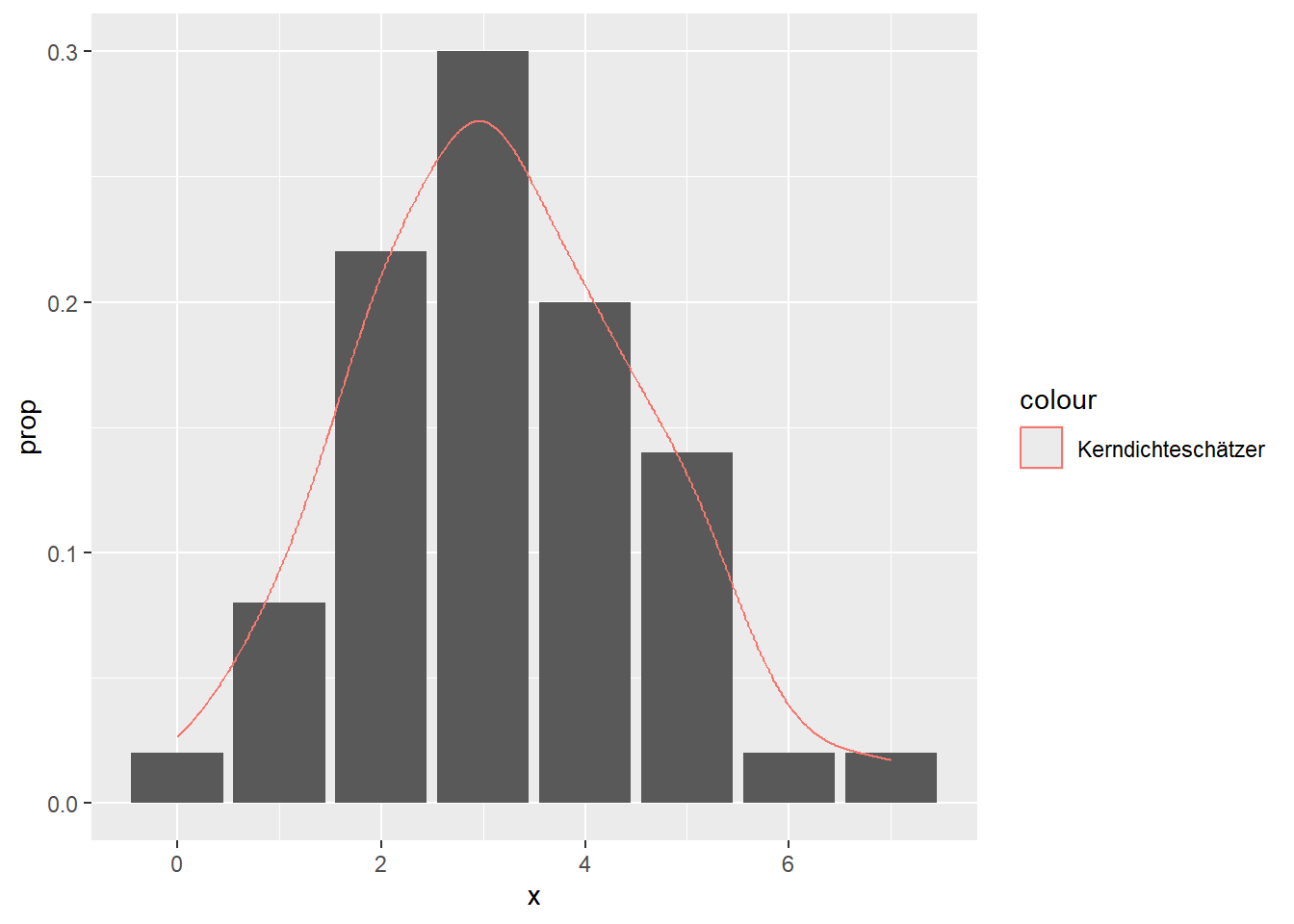
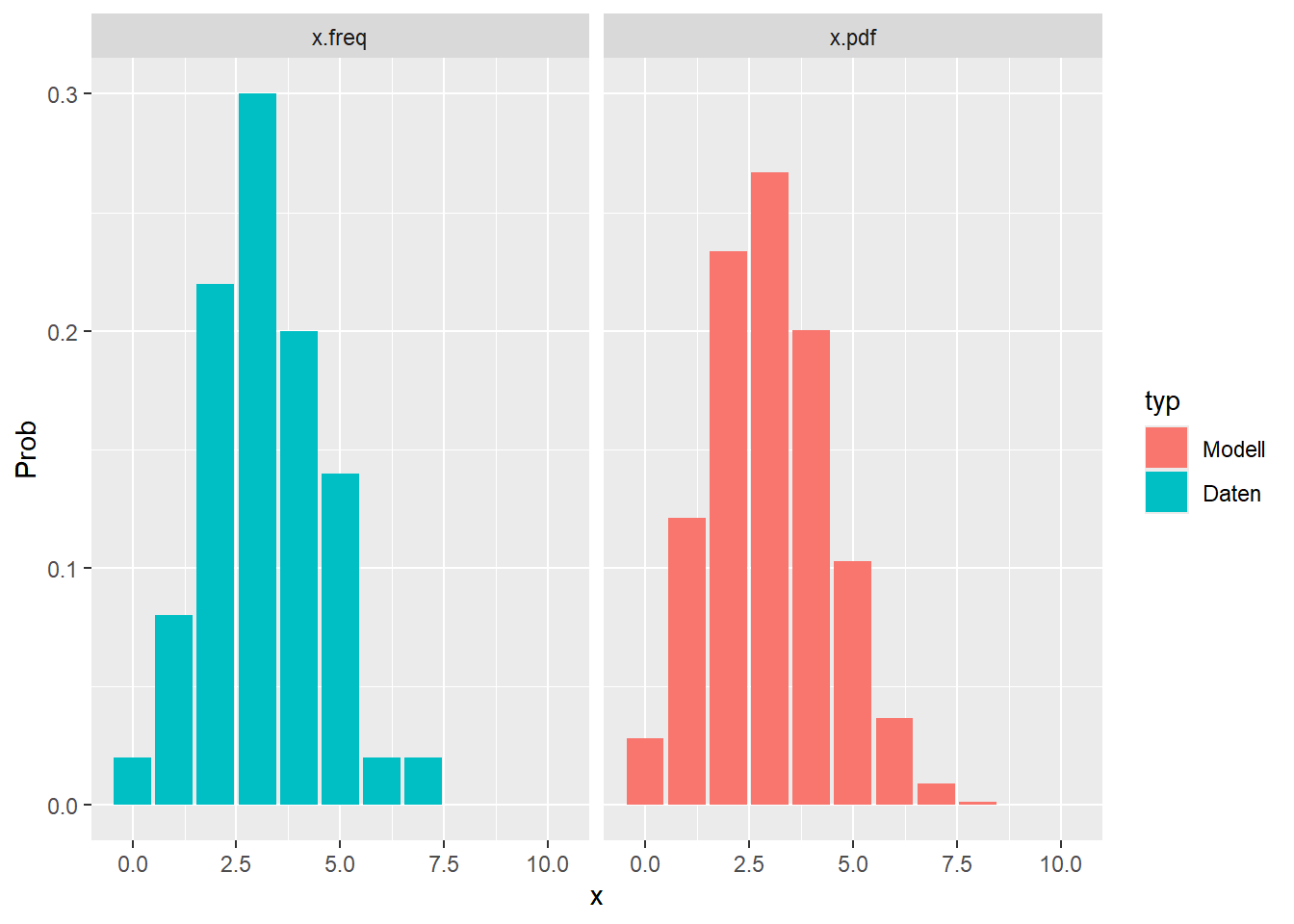
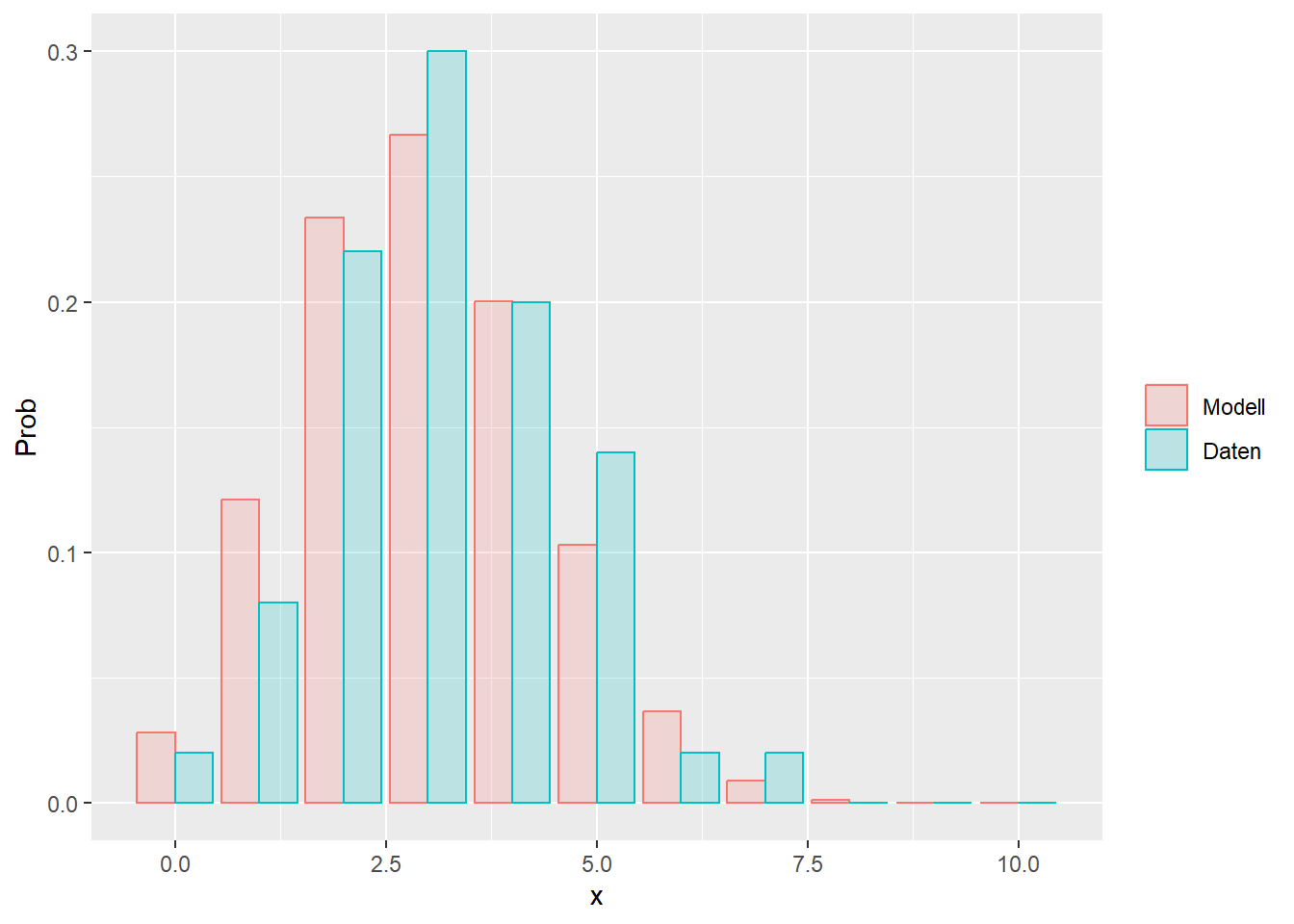
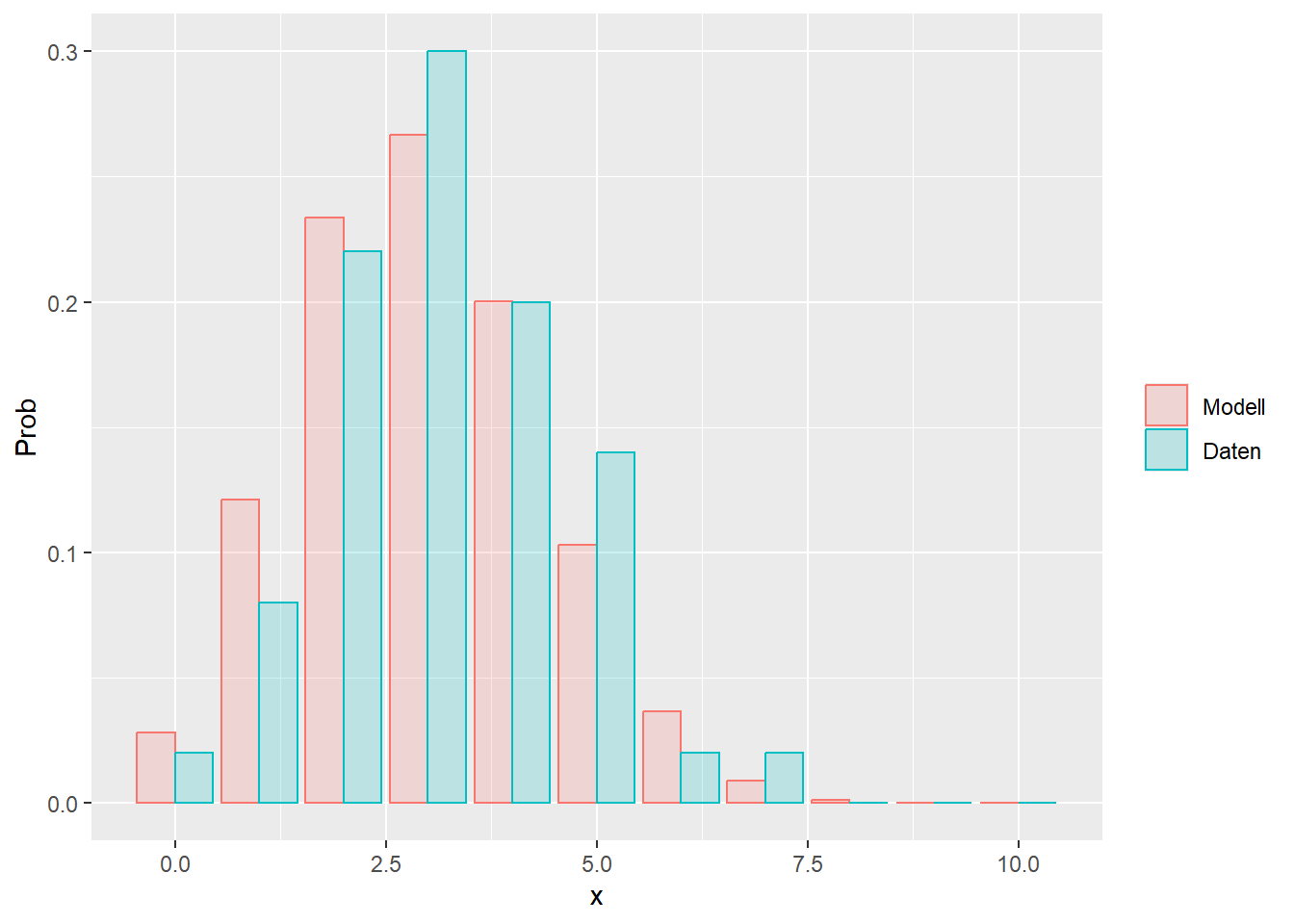
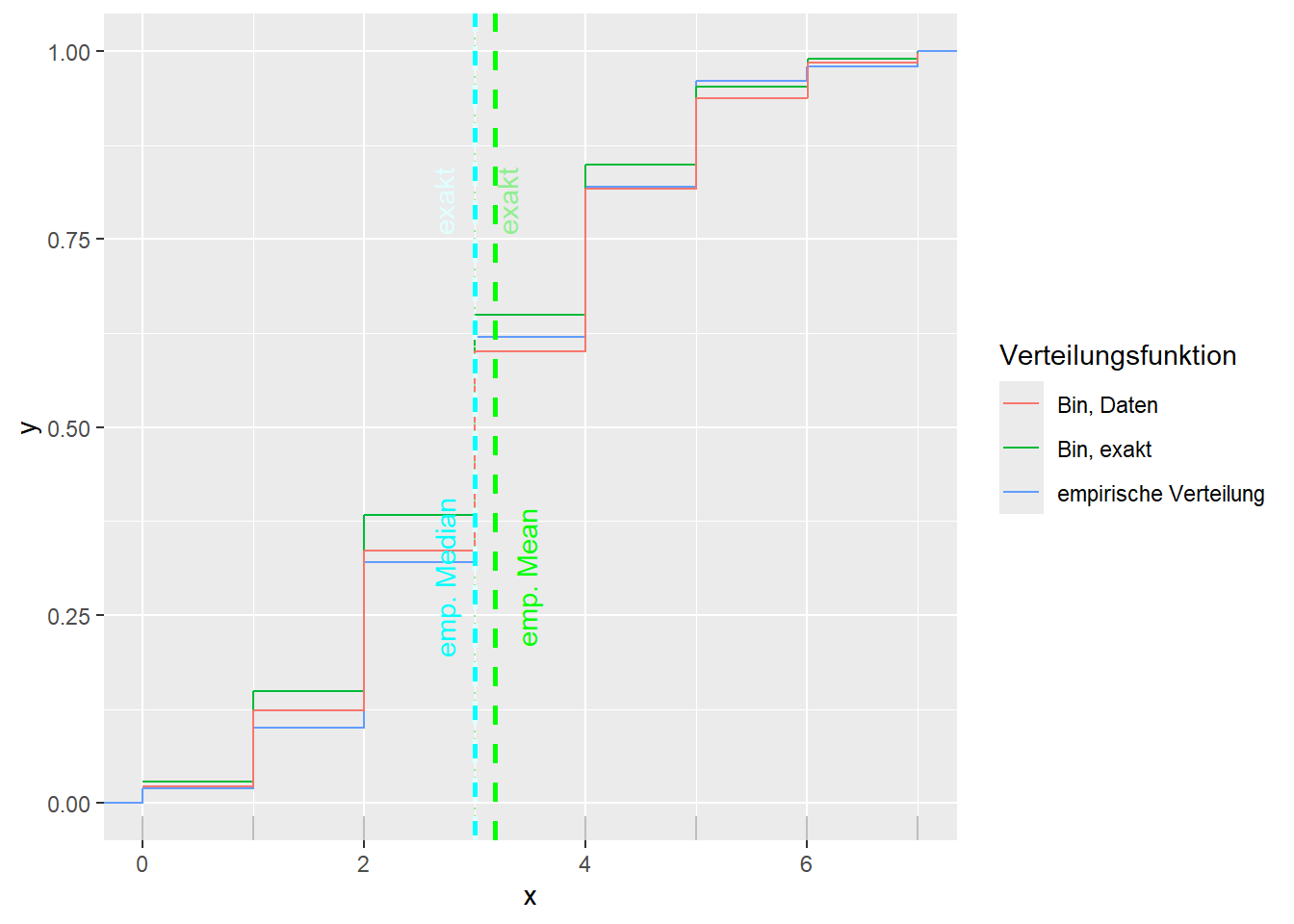
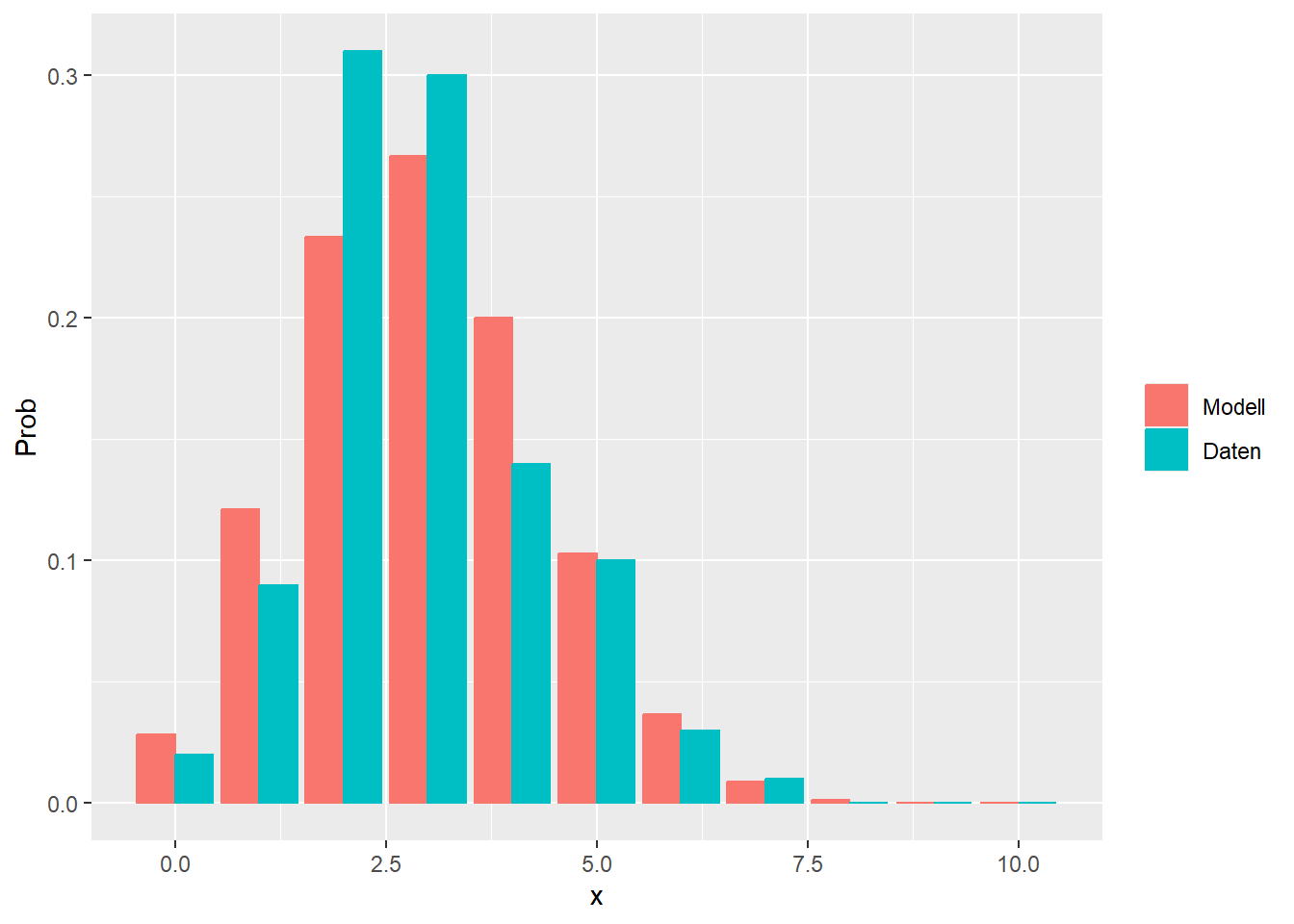
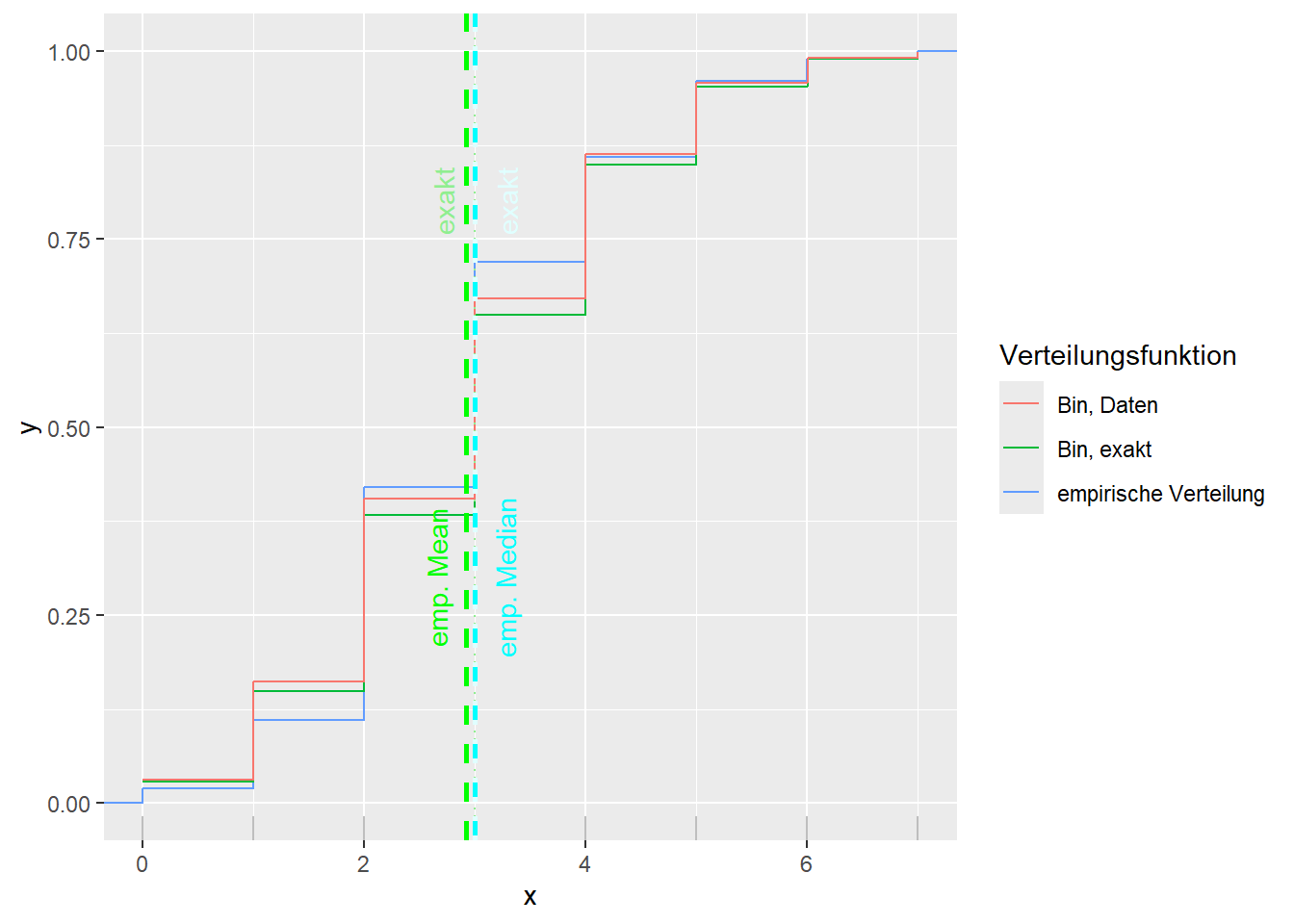
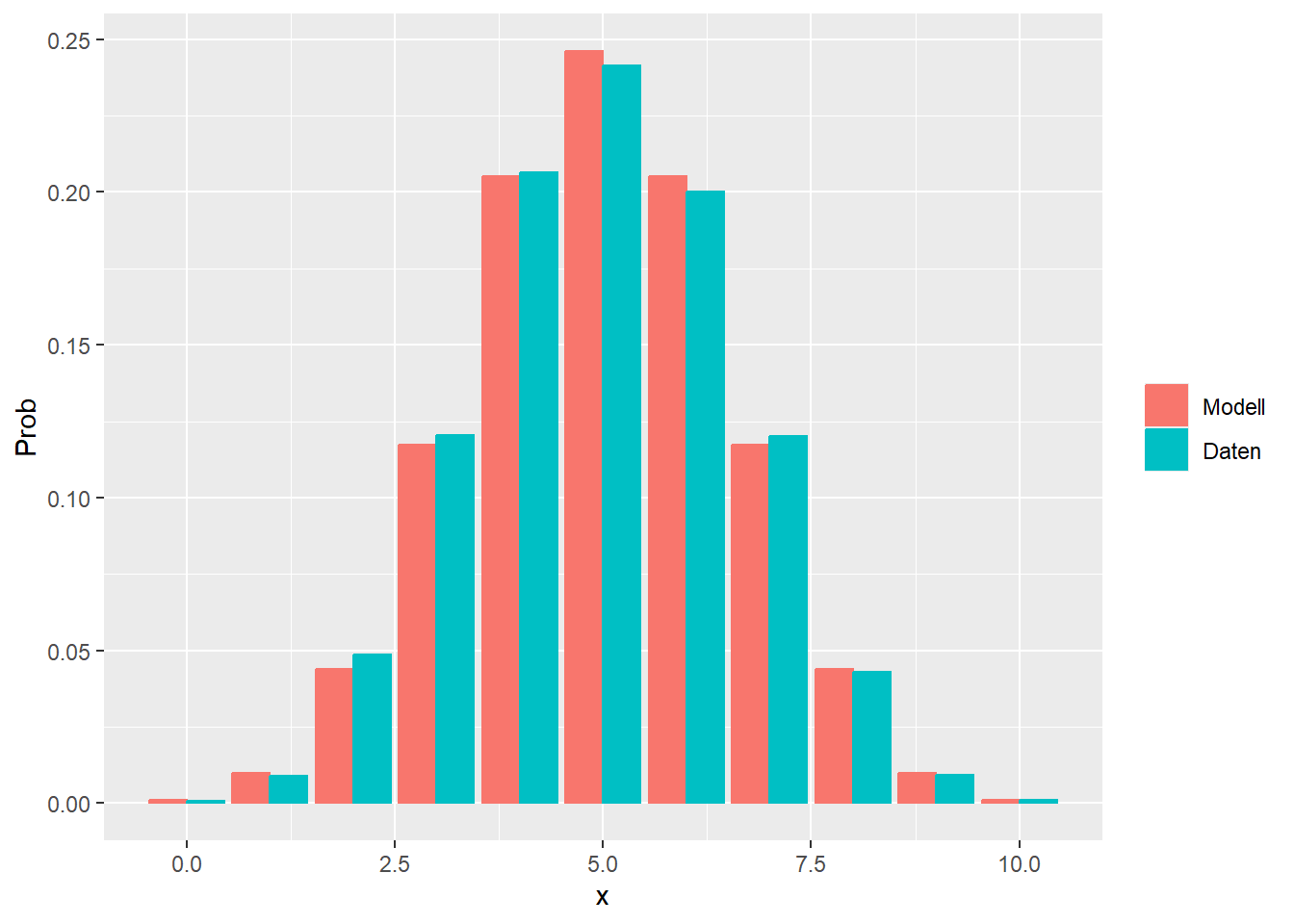
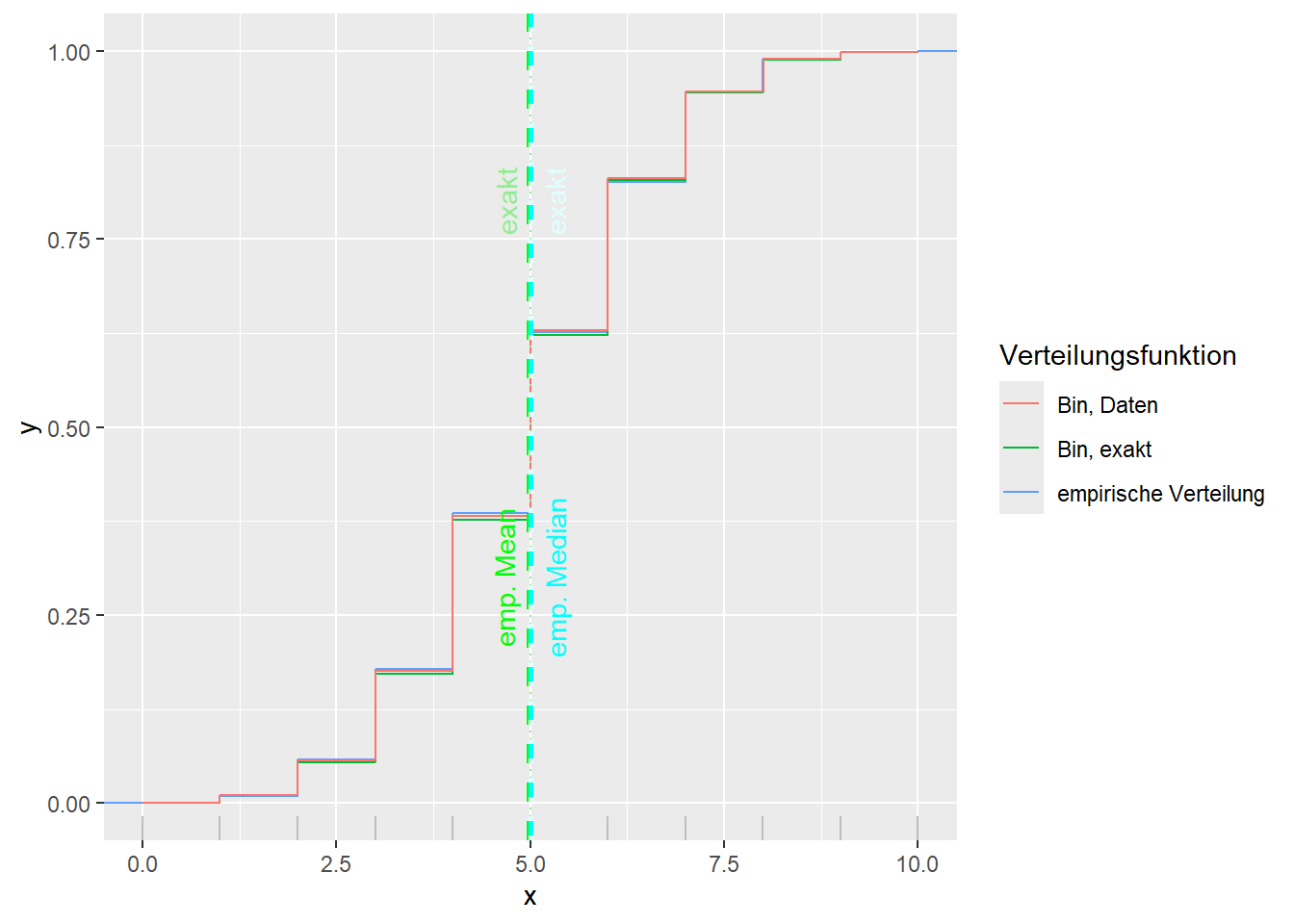
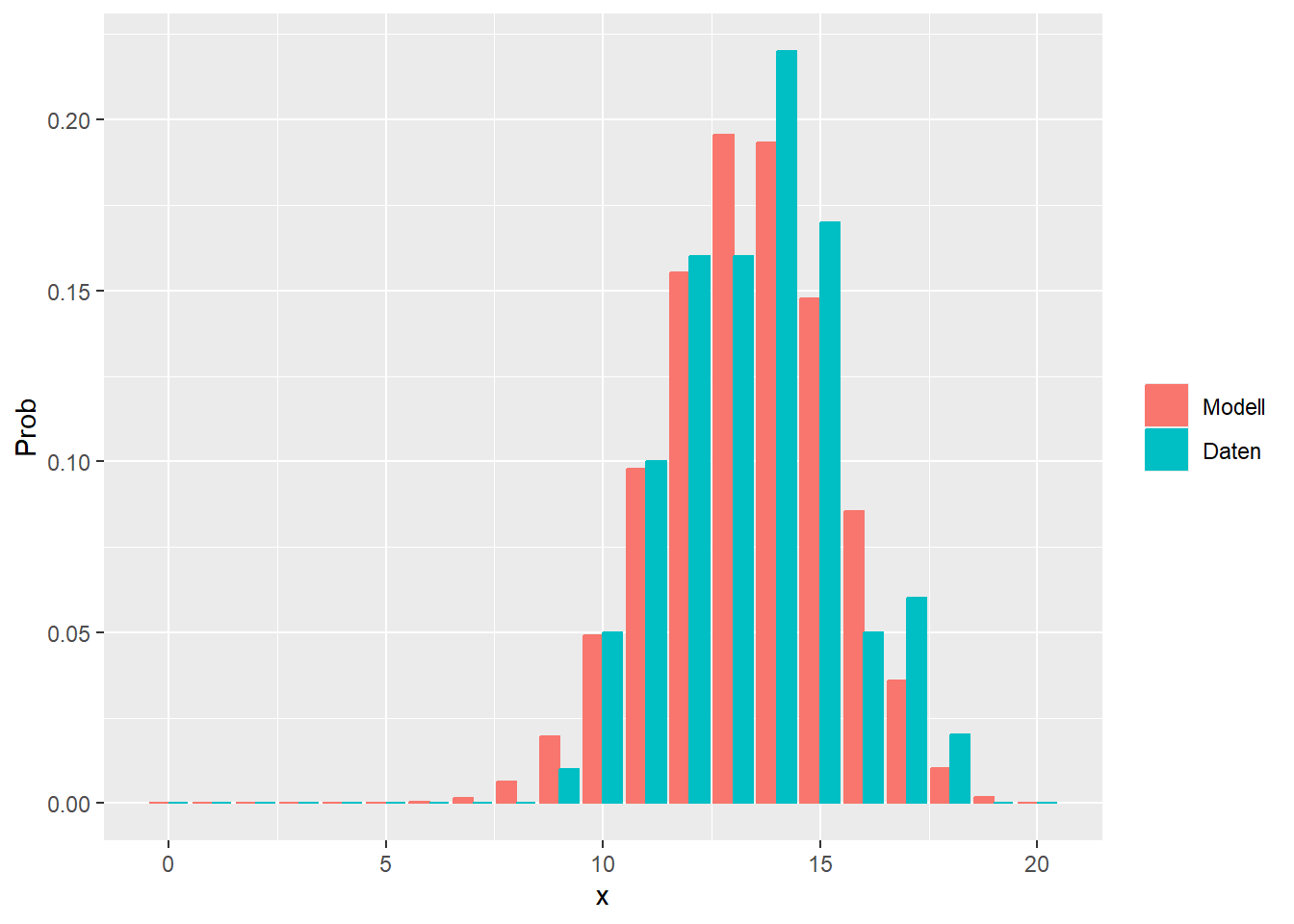
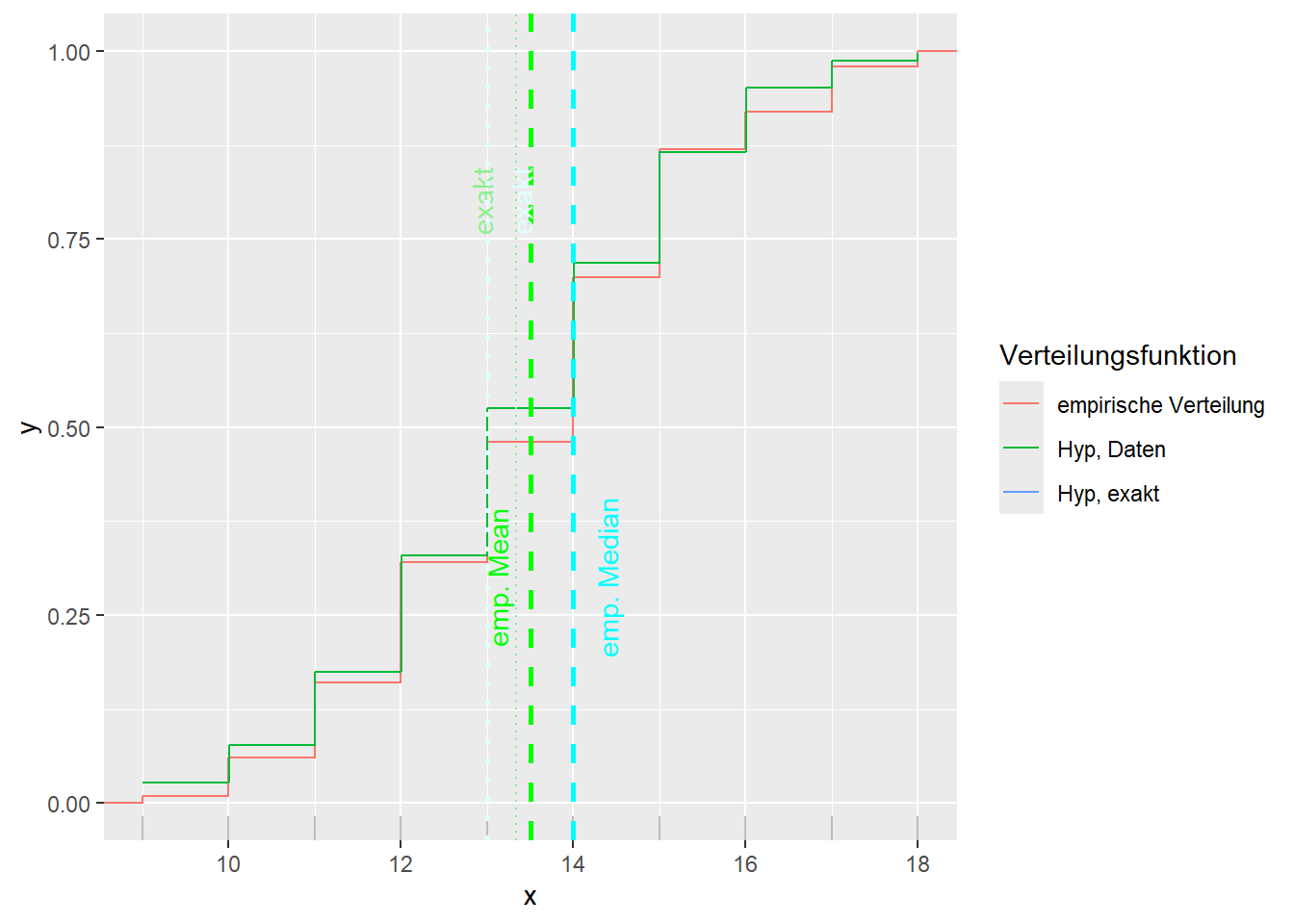
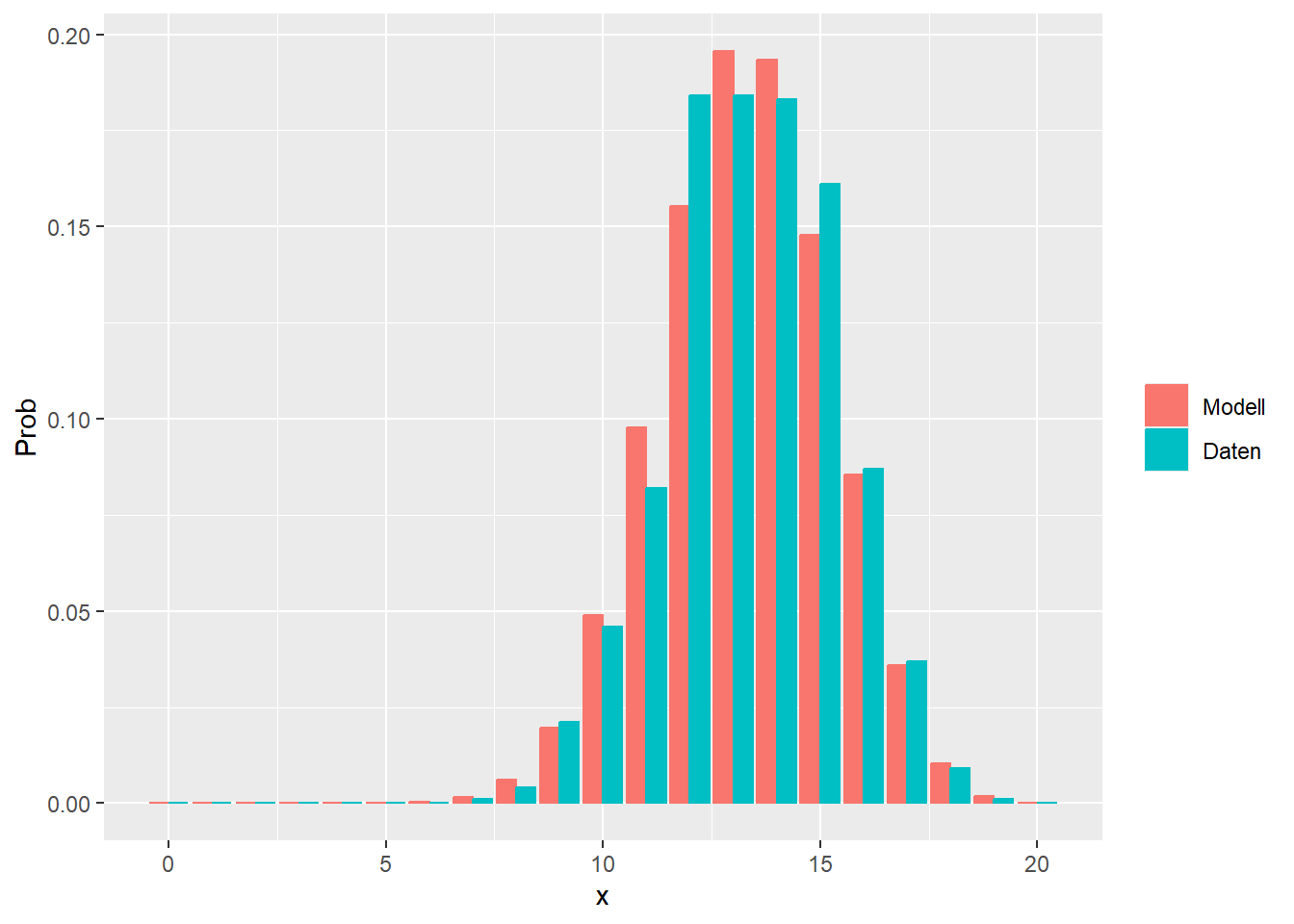
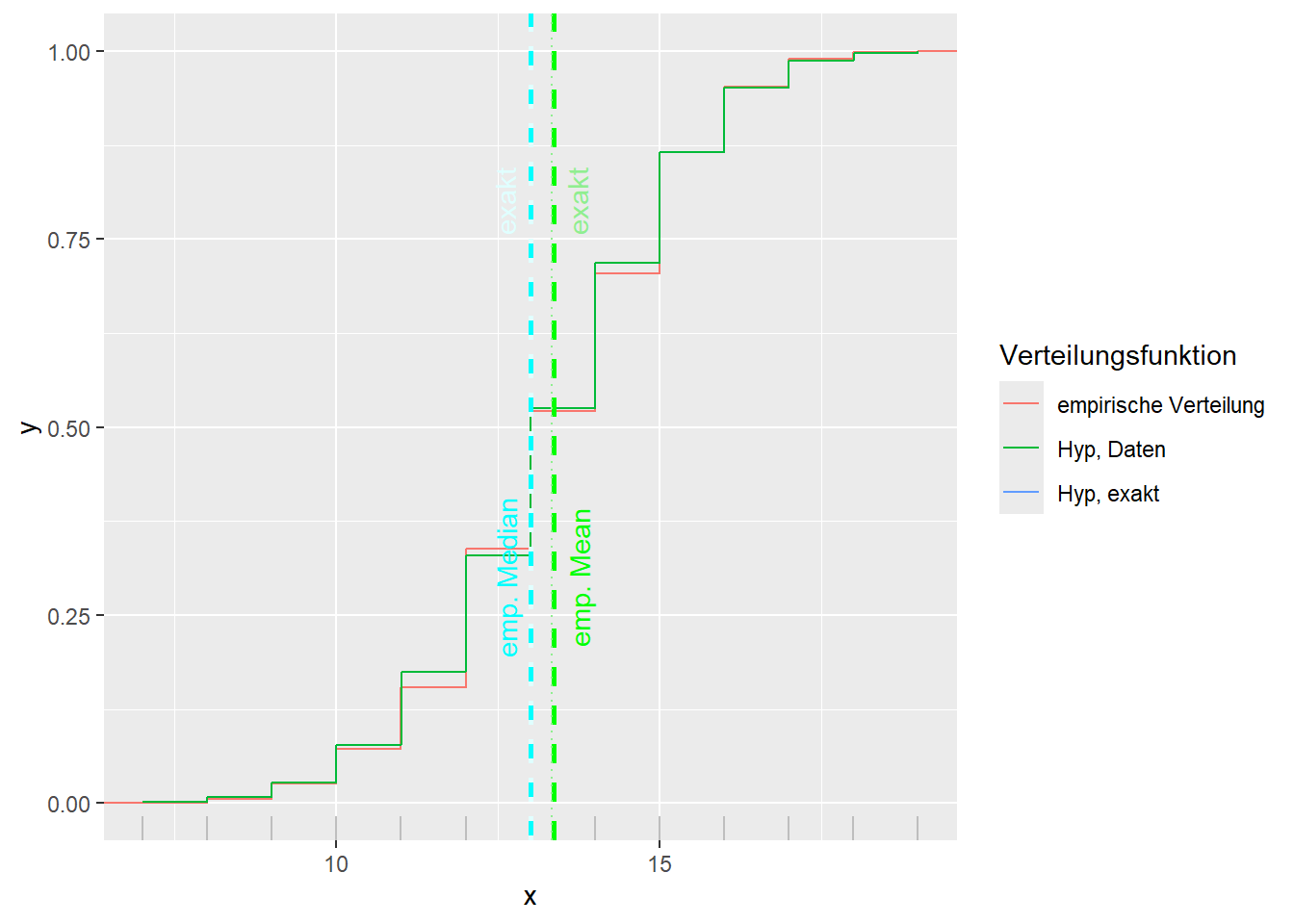
--- title: "Illustration von Verteilungen" number-sections: true author: - name: Dana Uhlig email: dana.uhlig@math.tu-chemnitz.de url: https://www.tu-chemnitz.de/mathematik/fima/dana/ #affiliation: TU Chemnitz format: html: code-fold: show code-summary: "R-Code" code-copy: true code-tools: true code-overflow: wrap toc: true toc-location: left toc-expand: true editor: visual --- ```{r setup, include=FALSE} knitr::opts_chunk$set(echo = TRUE, tidy='styler') ``` # Vorbereitung: Arbeiten mit Statistiksoftware R <https://www.r-project.org/> Empfehlung Editor: RStudio auf <https://posit.co/products/open-source/rstudio/> ```{r} = c ("tidyverse" , "latex2exp" )= wants %in% rownames (installed.packages ())if (any (! has)) install.packages (wants[! has])#fuer schoene Grafiken, Stichwort ggplot library ("tidyverse" )library (latex2exp)``` ```{r} = 50 ``` # Stetige Verteilungen ## Exponentialverteilung ```{r} = 2 exakt.mean = 1 / mu)exakt.var = 1 / (mu^ 2 ))exakt.median = log (2 )/ mu)#simulieren Daten nach Exponentialverteilung = rexp (n = n, rate = mu)= tibble (x)#empirische Masszahlen x.mean = mean (x))x.median = median (x))x.sd = sd (x))#nur fuer Graphiken = x.mean - x.median= - 0.2 * (d< 0 ) + 0.2 * (d>= 0 )= 0.2 * (d< 0 ) - 0.2 * (d>= 0 )``` ```{r} ggplot (data = x.data, aes (x)) + geom_histogram (aes (y= after_stat (density))) + #Histogramm der relativen Haeufigkeiten #geom_rug(aes(col="Daten")) + geom_rug (col= "gray" ) + #add rug plot: Daten auf der X-Achse geom_density (aes (col = "Kerndichteschätzer" )) + stat_function (fun = dexp, args = list (rate= mu), aes (col= "Exp, exakt" ))+ stat_function (fun = dexp, args = list (rate= 1 / x.mean), aes (col= "Exp, Daten" )) + #empirischer Mittelwert: geom_vline (aes (xintercept= x.mean),linetype= "dashed" , linewidth= 1 , colour= "green" ) + annotate ("text" , label = "emp. Mean" , x = x.mean + d.mean* x.sd, y = 0.3 , colour = "green" , angle = 90 ) + #exakter Mittelwert geom_vline (aes (xintercept= exakt.mean),linetype= "dotted" , linewidth= 0.5 , colour= "lightgreen" ) + annotate ("text" , label = "exakt" , x = exakt.mean + d.mean* sqrt (exakt.var), y = 1.9 , colour = "lightgreen" , angle = 90 ) + #empirischer Median geom_vline (aes (xintercept= x.median),linetype= "dashed" , linewidth= 1 , colour= "cyan" ) + annotate ("text" , label = "emp. Median" , x = x.median + d.median* x.sd, y = 0.3 , colour = "cyan" , angle = 90 ) + #exakter Median geom_vline (aes (xintercept= exakt.median),linetype= "dotted" , linewidth= 1 , colour= "lightcyan" ) + annotate ("text" , label = "exakt" , x = exakt.median + d.median* sqrt (exakt.var), y = 1.9 , colour = "lightcyan" , angle = 90 ) + labs (col = 'Dichte' )``` ```{r} ggplot (data = x.data, aes (x)) + stat_ecdf (aes (col = "empirische Verteilung" )) + geom_rug (col= "gray" ) + #add rug plot: Daten auf der X-Achse stat_function (fun = pexp, args = list (rate= mu), aes (col= "Exp, exakt" )) + stat_function (fun = pexp, args = list (rate= 1 / x.mean), aes (col= "Exp, Daten" )) + labs (col = 'Verteilungsfunktion' ) + #empirischer Mittelwert: geom_vline (aes (xintercept= x.mean),linetype= "dashed" , linewidth= 1 , colour= "green" ) + annotate ("text" , label = "emp. Mean" , x = x.mean + d.mean* x.sd, y = 0.3 , colour = "green" , angle = 90 ) + #exakter Mittelwert geom_vline (aes (xintercept= exakt.mean),linetype= "dotted" , linewidth= 0.5 , colour= "lightgreen" ) + annotate ("text" , label = "exakt" , x = exakt.mean + d.mean* sqrt (exakt.var), y = 0.8 , colour = "lightgreen" , angle = 90 ) + #empirischer Median geom_vline (aes (xintercept= x.median),linetype= "dashed" , linewidth= 1 , colour= "cyan" ) + annotate ("text" , label = "emp. Median" , x = x.median + d.median* x.sd, y = 0.3 , colour = "cyan" , angle = 90 ) + #exakter Median geom_vline (aes (xintercept= exakt.median),linetype= "dotted" , linewidth= 1 , colour= "lightcyan" ) + annotate ("text" , label = "exakt" , x = exakt.median + d.median* sqrt (exakt.var), y = 0.8 , colour = "lightcyan" , angle = 90 ) ``` ```{r} = function (n = 50 , mu = 2.0 ){= 1 / mu= 1 / (mu^ 2 )= log (2 )/ mu#simulieren Daten nach Exponentialverteilung = rexp (n = n, rate = mu)= tibble (x)#empirische Masszahlen = mean (x)= median (x)= sd (x)#nur fuer Graphiken = x.mean - x.median= - 0.2 * (d< 0 ) + 0.2 * (d>= 0 )= 0.2 * (d< 0 ) - 0.2 * (d>= 0 )= mu= ggplot (data = x.data, aes (x)) + geom_histogram (aes (y= after_stat (density))) + #Histogramm der relativen Haeufigkeiten #geom_rug(aes(col="Daten")) + geom_rug (col= "gray" ) + #add rug plot: Daten auf der X-Achse geom_density (aes (col = "Kerndichteschätzer" )) + stat_function (fun = dexp, args = list (rate= mu), aes (col= "Exp, exakt" ))+ stat_function (fun = dexp, args = list (rate= 1 / x.mean), aes (col= "Exp, Daten" )) + #empirischer Mittelwert: geom_vline (aes (xintercept= x.mean),linetype= "dashed" , linewidth= 1 , colour= "green" ) + annotate ("text" , label = "emp. Mean" , x = x.mean + d.mean* x.sd, y = 0.3 * max.dens, colour = "green" , angle = 90 ) + #exakter Mittelwert geom_vline (aes (xintercept= exakt.mean),linetype= "dotted" , linewidth= 0.5 , colour= "lightgreen" ) + annotate ("text" , label = "exakt" , x = exakt.mean + d.mean* sqrt (exakt.var), y = 1.0 * max.dens, colour = "lightgreen" , angle = 90 ) + #empirischer Median geom_vline (aes (xintercept= x.median),linetype= "dashed" , linewidth= 1 , colour= "cyan" ) + annotate ("text" , label = "emp. Median" , x = x.median + d.median* x.sd, y = 0.3 * max.dens, colour = "cyan" , angle = 90 ) + #exakter Median geom_vline (aes (xintercept= exakt.median),linetype= "dotted" , linewidth= 1 , colour= "lightcyan" ) + annotate ("text" , label = "exakt" , x = exakt.median + d.median* sqrt (exakt.var), y = 1.0 * max.dens, colour = "lightcyan" , angle = 90 ) + labs (col = 'Dichte' )print (graph.pdf)= ggplot (data = x.data, aes (x)) + stat_ecdf (aes (col = "empirische Verteilung" )) + geom_rug (col= "gray" ) + #add rug plot: Daten auf der X-Achse stat_function (fun = pexp, args = list (rate= mu), aes (col= "Exp, exakt" )) + stat_function (fun = pexp, args = list (rate= 1 / x.mean), aes (col= "Exp, Daten" )) + labs (col = 'Verteilungsfunktion' ) + #empirischer Mittelwert: geom_vline (aes (xintercept= x.mean),linetype= "dashed" , linewidth= 1 , colour= "green" ) + annotate ("text" , label = "emp. Mean" , x = x.mean + d.mean* x.sd, y = 0.3 , colour = "green" , angle = 90 ) + #exakter Mittelwert geom_vline (aes (xintercept= exakt.mean),linetype= "dotted" , linewidth= 0.5 , colour= "lightgreen" ) + annotate ("text" , label = "exakt" , x = exakt.mean + d.mean* sqrt (exakt.var), y = 0.8 , colour = "lightgreen" , angle = 90 ) + #empirischer Median geom_vline (aes (xintercept= x.median),linetype= "dashed" , linewidth= 1 , colour= "cyan" ) + annotate ("text" , label = "emp. Median" , x = x.median + d.median* x.sd, y = 0.3 , colour = "cyan" , angle = 90 ) + #exakter Median geom_vline (aes (xintercept= exakt.median),linetype= "dotted" , linewidth= 1 , colour= "lightcyan" ) + annotate ("text" , label = "exakt" , x = exakt.median + d.median* sqrt (exakt.var), y = 0.8 , colour = "lightcyan" , angle = 90 ) print (graph.cdf)#return(list(graph.pdf, graph.cdf)) ``` ```{r} sim_exp (n = 100 , mu = 2 )``` ```{r} sim_exp (n = 10000 , mu = 2 )``` ## Normalverteilung ```{r} = function (n = 50 , mu = 0.0 , sigma = 1.0 ){= mu= sigma^ 2 = qnorm (p = 0.5 , mean = mu, sd = sigma)#simulieren Daten nach Normalverteilung = rnorm (n = n, mean = mu, sd = sigma)= tibble (x)#empirische Masszahlen = mean (x)= median (x)= sd (x)#nur fuer Graphiken = x.mean - x.median= - 0.2 * (d< 0 ) + 0.2 * (d>= 0 )= 0.2 * (d< 0 ) - 0.2 * (d>= 0 )= 1 / (sqrt (2 * pi)* sigma)= ggplot (data = x.data, aes (x)) + geom_histogram (aes (y= after_stat (density))) + #Histogramm der relativen Haeufigkeiten #geom_rug(aes(col="Daten")) + geom_rug (col= "gray" ) + #add rug plot: Daten auf der X-Achse geom_density (aes (col = "Kerndichteschätzer" )) + stat_function (fun = dnorm, args = list (mean= mu, sd = sigma), aes (col= "NV, exakt" ))+ stat_function (fun = dnorm, args = list (mean= x.mean, sd = x.sd), aes (col= "NV, Daten" )) + #empirischer Mittelwert: geom_vline (aes (xintercept= x.mean),linetype= "dashed" , linewidth= 1 , colour= "green" ) + annotate ("text" , label = "emp. Mean" , x = x.mean + d.mean* x.sd, y = 0.3 * max.dens, colour = "green" , angle = 90 ) + #exakter Mittelwert geom_vline (aes (xintercept= exakt.mean),linetype= "dotted" , linewidth= 0.5 , colour= "lightgreen" ) + annotate ("text" , label = "exakt" , x = exakt.mean + d.mean* sqrt (exakt.var), y = 0.9 * max.dens, colour = "lightgreen" , angle = 90 ) + #empirischer Median geom_vline (aes (xintercept= x.median),linetype= "dashed" , linewidth= 1 , colour= "cyan" ) + annotate ("text" , label = "emp. Median" , x = x.median + d.median* x.sd, y = 0.3 * max.dens, colour = "cyan" , angle = 90 ) + #exakter Median geom_vline (aes (xintercept= exakt.median),linetype= "dotted" , linewidth= 1 , colour= "lightcyan" ) + annotate ("text" , label = "exakt" , x = exakt.median + d.median* sqrt (exakt.var), y = 0.9 * max.dens, colour = "lightcyan" , angle = 90 ) + labs (col = 'Dichte' )print (graph.pdf)= ggplot (data = x.data, aes (x)) + stat_ecdf (aes (col = "empirische Verteilung" )) + geom_rug (col= "gray" ) + #add rug plot: Daten auf der X-Achse stat_function (fun = pnorm, args = list (mean= mu, sd = sigma), aes (col= "NV, exakt" )) + stat_function (fun = pnorm, args = list (mean= mu, sd = sigma), aes (col= "NV, Daten" )) + labs (col = 'Verteilungsfunktion' ) + #empirischer Mittelwert: geom_vline (aes (xintercept= x.mean),linetype= "dashed" , linewidth= 1 , colour= "green" ) + annotate ("text" , label = "emp. Mean" , x = x.mean + d.mean* x.sd, y = 0.3 , colour = "green" , angle = 90 ) + #exakter Mittelwert geom_vline (aes (xintercept= exakt.mean),linetype= "dotted" , linewidth= 0.5 , colour= "lightgreen" ) + annotate ("text" , label = "exakt" , x = exakt.mean + d.mean* sqrt (exakt.var), y = 0.8 , colour = "lightgreen" , angle = 90 ) + #empirischer Median geom_vline (aes (xintercept= x.median),linetype= "dashed" , linewidth= 1 , colour= "cyan" ) + annotate ("text" , label = "emp. Median" , x = x.median + d.median* x.sd, y = 0.3 , colour = "cyan" , angle = 90 ) + #exakter Median geom_vline (aes (xintercept= exakt.median),linetype= "dotted" , linewidth= 1 , colour= "lightcyan" ) + annotate ("text" , label = "exakt" , x = exakt.median + d.median* sqrt (exakt.var), y = 0.8 , colour = "lightcyan" , angle = 90 ) print (graph.cdf)#return(list(graph.pdf, graph.cdf)) ``` ```{r} sim_normal (n = 100 , mu = 3 , sigma = 2 )``` ```{r} sim_normal (n = 10000 , mu = 3 , sigma = 2 )``` # Diskrete Verteilungen ## Binomialverteilung ```{r} = 0.3 = 10 # bekannt exakt.mean = m* p)exakt.var = m* p* (1 - p))exakt.median = qbinom (p= 0.5 , size = m, prob = p))#simulieren Daten nach Binomialverteilung = rbinom (n = n, size = m, prob = p)= tibble (x)#empirische Masszahlen x.mean = mean (x))x.median = median (x))x.sd = sd (x))p.data = x.mean / m)#nur fuer Graphiken = x.mean - x.median= - 0.2 * (d< 0 ) + 0.2 * (d>= 0 )= 0.2 * (d< 0 ) - 0.2 * (d>= 0 )#absolute Hauefigkeiten table (x)#rel. Häufigkeiten table (x)/ length (x)#Wahrscheinlichkeiten = pbinom (q= 0 : m, size = m, prob = p)round (x.probs,digits = 3 )``` ```{r} ggplot (data = x.data, aes (x)) + geom_histogram (aes (y= after_stat (density))) + geom_density ()``` ```{r} ggplot (data = x.data, aes (x)) + geom_bar (aes (y= after_stat (prop))) + #geom_rug(col="gray") + #add rug plot: Daten auf der X-Achse geom_density (aes (col = "Kerndichteschätzer" )) ``` ```{r} = x.data %>% group_by (x) %>% summarise (n = n ()) %>% mutate (x.freq = n / sum (n))%>% table ()= dbinom (x = 0 : m, size = m, prob = p)#x.cdf = pbinom(q = 0:m, size = m, prob = p) #x.exakt = tibble(x=0:m, x.pdf, x.cdf) = tibble (x= 0 : m, x.pdf)= full_join (x.exakt, x.emp,by = "x" ) = pivot_longer (x.all, cols = c (x.pdf, x.freq), values_to = "Prob" , names_to = "type" ) %>% mutate (typ = recode_factor (type, x.pdf = "Modell" , x.freq = "Daten" )) %>% replace_na (list (Prob = 0 ))print (x.all.long, n = 2 * (m+ 1 ))``` ```{r} %>% group_by (typ) %>% summarise ( gesamt = sum (Prob))``` ```{r} ggplot (data = x.all.long, aes (x,y= Prob, fill = typ)) + geom_col () + facet_wrap (~ type)``` ```{r} ggplot (data = x.all.long, aes (x,y= Prob, col= typ, fill= typ)) + geom_col (alpha = 0.2 , position= "dodge" ) + labs (fill = NULL , col = NULL )``` ```{r} ggplot (data = x.all.long, aes (x,y= Prob, col= typ, fill= typ)) + geom_bar (stat = "identity" , alpha = 0.2 ,position= "dodge" ) + labs (fill = NULL , col = NULL )``` ```{r} ggplot (data = x.data, aes (x)) + stat_ecdf (aes (col = "empirische Verteilung" )) + geom_rug (col= "gray" ) + #add rug plot: Daten auf der X-Achse stat_function (fun = pbinom, args = list (size= m, prob = p), aes (col= "Bin, exakt" ), n = 2000 ) + stat_function (fun = pbinom, args = list (size= m, prob = p.data), aes (col= "Bin, Daten" ), n= 2000 ) + labs (col = 'Verteilungsfunktion' ) + #empirischer Mittelwert: geom_vline (aes (xintercept= x.mean),linetype= "dashed" , linewidth= 1 , colour= "green" ) + annotate ("text" , label = "emp. Mean" , x = x.mean + d.mean* x.sd, y = 0.3 , colour = "green" , angle = 90 ) + #exakter Mittelwert geom_vline (aes (xintercept= exakt.mean),linetype= "dotted" , linewidth= 0.5 , colour= "lightgreen" ) + annotate ("text" , label = "exakt" , x = exakt.mean + d.mean* sqrt (exakt.var), y = 0.8 , colour = "lightgreen" , angle = 90 ) + #empirischer Median geom_vline (aes (xintercept= x.median),linetype= "dashed" , linewidth= 1 , colour= "cyan" ) + annotate ("text" , label = "emp. Median" , x = x.median + d.median* x.sd, y = 0.3 , colour = "cyan" , angle = 90 ) + #exakter Median geom_vline (aes (xintercept= exakt.median),linetype= "dotted" , linewidth= 1 , colour= "lightcyan" ) + annotate ("text" , label = "exakt" , x = exakt.median + d.median* sqrt (exakt.var), y = 0.8 , colour = "lightcyan" , angle = 90 ) ``` ```{r} = function (n = 50 , m = 10 , p = 0.5 ){= m* p= m* p* (1 - p)= qbinom (p= 0.5 , size = m, prob = p)#simulieren Daten nach Binomialverteilung = rbinom (n = n, size = m, prob = p)= tibble (x)#empirische Masszahlen = mean (x)= median (x)= sd (x)= x.mean / m#nur fuer Graphiken = x.mean - x.median= - 0.2 * (d< 0 ) + 0.2 * (d>= 0 )= 0.2 * (d< 0 ) - 0.2 * (d>= 0 )= x.data %>% group_by (x) %>% summarise (n = n ()) %>% mutate (x.freq = n / sum (n))#x.data %>% table() = dbinom (x = 0 : m, size = m, prob = p)= tibble (x= 0 : m, x.pdf)= full_join (x.exakt, x.emp,by = "x" ) = pivot_longer (x.all, cols = c (x.pdf, x.freq), values_to = "Prob" , names_to = "type" ) %>% mutate (typ = recode_factor (type, x.pdf = "Modell" , x.freq = "Daten" )) %>% replace_na (list (Prob = 0 ))= ggplot (data = x.all.long, aes (x,y= Prob, col= typ, fill= typ)) + geom_bar (stat = "identity" , position= "dodge" ) + labs (fill = NULL , col = NULL )= ggplot (data = x.data, aes (x)) + stat_ecdf (aes (col = "empirische Verteilung" )) + geom_rug (col= "gray" ) + #add rug plot: Daten auf der X-Achse stat_function (fun = pbinom, args = list (size= m, prob = p), aes (col= "Bin, exakt" ), n = 2000 ) + stat_function (fun = pbinom, args = list (size= m, prob = p.data), aes (col= "Bin, Daten" ), n= 2000 ) + labs (col = 'Verteilungsfunktion' ) + #empirischer Mittelwert: geom_vline (aes (xintercept= x.mean),linetype= "dashed" , linewidth= 1 , colour= "green" ) + annotate ("text" , label = "emp. Mean" , x = x.mean + d.mean* x.sd, y = 0.3 , colour = "green" , angle = 90 ) + #exakter Mittelwert geom_vline (aes (xintercept= exakt.mean),linetype= "dotted" , linewidth= 0.5 , colour= "lightgreen" ) + annotate ("text" , label = "exakt" , x = exakt.mean + d.mean* sqrt (exakt.var), y = 0.8 , colour = "lightgreen" , angle = 90 ) + #empirischer Median geom_vline (aes (xintercept= x.median),linetype= "dashed" , linewidth= 1 , colour= "cyan" ) + annotate ("text" , label = "emp. Median" , x = x.median + d.median* x.sd, y = 0.3 , colour = "cyan" , angle = 90 ) + #exakter Median geom_vline (aes (xintercept= exakt.median),linetype= "dotted" , linewidth= 1 , colour= "lightcyan" ) + annotate ("text" , label = "exakt" , x = exakt.median + d.median* sqrt (exakt.var), y = 0.8 , colour = "lightcyan" , angle = 90 ) print (graph.pdf)print (graph.cdf)#return(list(graph.pdf,graph.cdf)) ``` ```{r} sim_binomial (n = 100 , m = 10 , p = 0.3 )``` ```{r} sim_binomial (n = 10000 , m = 10 , p = 0.5 )``` ## Hypergeometrische Verteilung ```{r} = function (n = 50 , White = 10 , Black = 10 , k = 10 ){#X = number white balls by k x drawing without replacement = White/ (White + Black)= k* p= k * p* (1 - p)* (White + Black - k)/ (White + Black - 1 )= qhyper (p= 0.5 , m = White, n = Black, k = k)#simulieren Daten nach Binomialverteilung = rhyper (nn = n, m = White, n = Black, k = k)= tibble (x)#empirische Masszahlen = mean (x)= median (x)= sd (x)#nur fuer Graphiken = x.mean - x.median= - 0.2 * (d< 0 ) + 0.2 * (d>= 0 )= 0.2 * (d< 0 ) - 0.2 * (d>= 0 )= x.data %>% group_by (x) %>% summarise (n = n ()) %>% mutate (x.freq = n / sum (n))#x.data %>% table() = dhyper (x = 0 : k, m = White, n = Black, k = k)= tibble (x= 0 : k, x.pdf)= full_join (x.exakt, x.emp,by = "x" ) = pivot_longer (x.all, cols = c (x.pdf, x.freq), values_to = "Prob" , names_to = "type" ) %>% mutate (typ = recode_factor (type, x.pdf = "Modell" , x.freq = "Daten" )) %>% replace_na (list (Prob = 0 ))= ggplot (data = x.all.long, aes (x,y= Prob, col= typ, fill= typ)) + geom_bar (stat = "identity" , position= "dodge" ) + labs (fill = NULL , col = NULL )= ggplot (data = x.data, aes (x)) + stat_ecdf (aes (col = "empirische Verteilung" )) + geom_rug (col= "gray" ) + #add rug plot: Daten auf der X-Achse stat_function (fun = phyper, args = list (m = White, n = Black, k = k), aes (col= "Hyp, exakt" ), n = 2000 ) + stat_function (fun = phyper, args = list (m = White, n = Black, k = k), aes (col= "Hyp, Daten" ), n= 2000 ) + labs (col = 'Verteilungsfunktion' ) + #empirischer Mittelwert: geom_vline (aes (xintercept= x.mean),linetype= "dashed" , linewidth= 1 , colour= "green" ) + annotate ("text" , label = "emp. Mean" , x = x.mean + d.mean* x.sd, y = 0.3 , colour = "green" , angle = 90 ) + #exakter Mittelwert geom_vline (aes (xintercept= exakt.mean),linetype= "dotted" , linewidth= 0.5 , colour= "lightgreen" ) + annotate ("text" , label = "exakt" , x = exakt.mean + d.mean* sqrt (exakt.var), y = 0.8 , colour = "lightgreen" , angle = 90 ) + #empirischer Median geom_vline (aes (xintercept= x.median),linetype= "dashed" , linewidth= 1 , colour= "cyan" ) + annotate ("text" , label = "emp. Median" , x = x.median + d.median* x.sd, y = 0.3 , colour = "cyan" , angle = 90 ) + #exakter Median geom_vline (aes (xintercept= exakt.median),linetype= "dotted" , linewidth= 1 , colour= "lightcyan" ) + annotate ("text" , label = "exakt" , x = exakt.median + d.median* sqrt (exakt.var), y = 0.8 , colour = "lightcyan" , angle = 90 ) print (graph.pdf)print (graph.cdf)#return(list(graph.pdf,graph.cdf)) ``` ```{r} sim_hyper (n = 100 , White = 100 , Black = 50 , k = 20 )``` ```{r} sim_hyper (n = 1000 , White = 100 , Black = 50 , k = 20 )```